Genetic diversity of chilean and brazilian alstroemeria species assessed by aflp analysis
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT One to three accessions of 22 _Alstroemeria_ species, an interspecific hybrid (_A. aurea_ × _A. inodora_), and single accessions of _Bomarea salsilla_ and _Leontochir ovallei_ were
evaluated using the AFLP-marker technique to estimate the genetic diversity within the genus _Alstroemeria_. Three primer combinations generated 716 markers and discriminated all
_Alstroemeria_ species. The dendrogram inferred from the AFLP fingerprints supported the conjecture of the generic separation of the Chilean and Brazilian _Alstroemeria_ species. The
principal co-ordinate plot showed the separate allocation of the _A. ligtu_ group and the allocation of _A. aurea_, which has a wide range of geographical distribution and genetic variation,
in the middle of other _Alstroemeria_ species. The genetic distances, based on AFLP markers, determined the genomic contribution of the parents to the interspecific hybrid. SIMILAR CONTENT
BEING VIEWED BY OTHERS DNA FINGERPRINTING, FIXATION-INDEX (FST), AND ADMIXTURE MAPPING OF SELECTED BAMBARA GROUNDNUT (_VIGNA SUBTERRANEA_ [L.] VERDC.) ACCESSIONS USING ISSR MARKERS SYSTEM
Article Open access 15 July 2021 INTER SIMPLE SEQUENCE REPEAT MARKERS TO ASSESS GENETIC DIVERSITY OF THE DESERT DATE (_BALANITES AEGYPTIACA_ DEL.) FOR SAHELIAN ECOSYSTEM RESTORATION Article
Open access 11 September 2020 A SET OF SSR MARKERS TO CHARACTERIZE GENETIC DIVERSITY IN ALL _VIBURNUM_ SPECIES Article Open access 01 April 2023 INTRODUCTION The genus _Alstroemeria_
includes approximately 60 described species of rhizomatous, herbaceous plants, with Chile and Brazil as the main centres of diversity (Uphof, 1952; Bayer, 1987; Aker & Healy, 1990). The
Chilean and Brazilian _Alstroemeria_ are recognized as representatives of different branches of the genus. The family of Alstroemeriaceae, to which _Alstroemeria_ belongs, includes several
related genera, such as _Bomarea_ Mirbel, the monotype _Leontochir ovallei_ Phil. and _Schickendantzia_ Pax (Dahlgren & Clifford, 1982; Hutchinson, 1973). The species classification in
_Alstroemeria_ is based on an evaluation of morphological traits of the flower, stem, leaf, fruit and rhizome (Bayer, 1987). The available biosystematic information on _Alstroemeria_ species
is restricted to the Chilean species, as described in the monograph of Bayer (1987). Little is known about the classification of the Brazilian species (Meerow & Tombolato, 1996).
Furthermore, morphology-based identification is rather difficult because morphological characteristics can vary considerably in different environmental conditions (Bayer, 1987). The immense
genetic variation present in the genus _Alstroemeria_ offers many opportunities for the improvement and renewal of cultivars. Therefore, identification of genetic relationships at the
species level could be very useful for breeding in supporting the selection of crossing combinations from large sets of parental genotypes, thus broadening the genetic basis of breeding
programmes (Frei et al., 1986). The species used in the study reported here are commonly used in the breeding programme of _Alstroemeria_ for cut flowers and pot plants. Molecular techniques
have become increasingly significant for biosystematic studies (Soltis et al., 1992). RAPD markers were used for the identification of genetic relationships between _Alstroemeria_ species
and cultivars (Anastassopoulos & Keil, 1996; Dubouzet et al., 1997; Picton & Hughes, 1997). In recent years a novel PCR-based marker technique, AFLP (Vos et al., 1995), has been
developed and used for genetic studies in numerous plants including lettuce (Hill et al., 1996), lentil (Sharma et al., 1996), bean (Tohme et al., 1996), tea (Paul et al., 1997), barley
(Schut et al., 1997), and wild potato species (Kardolus et al., 1998). These studies indicated that AFLP is highly applicable for molecular discrimination at the species level. The technique
has also been optimized for use in species such as _Alstroemeria_ spp., which are characterized by a large genome size (2C-value: 37–79 pg) (Han et al., 1999). In this study, we produced
AFLP fingerprints of 22 _Alstroemeria_ species, one interspecific hybrid (_A. aurea × A. inodora_) and the distantly related species _Bomarea salsilla_ and _Leontochir ovallei_, and we
analysed their genetic relationships. The interspecific hybrid was included in our study in order to investigate the possibility of identifying the parental genotypes. MATERIALS AND METHODS
PLANT MATERIAL Seeds and plants of 22 _Alstroemeria_ species were obtained from botanical gardens and commercial breeders. The collection has been maintained for many years in the greenhouse
of Unifarm at the Wageningen Agricultural University. When available, three accessions were selected for each _Alstroemeria_ species, and both _B. salsilla_ and _L. ovallei_ were chosen as
outgroups. One interspecific hybrid (_A. aurea_ × _A. inodora_) was obtained from earlier research (Buitendijk et al., 1995) (Table 1). All accessions were identified according to their
morphological traits (Uphof, 1952; Bayer, 1987). AFLP PROTOCOL Genomic DNA was isolated from young leaves of greenhouse-grown plants using the cetyltrimethy-lammonium bromide (CTAB) method
according to Rogers & Bendich (1988). The AFLP technique followed the method of Vos et al. (1995) with modifications of selective bases of pre- and final amplifications (Han et al.,
1999). To assess interspecific variation, autoradiograms comprising the AFLP fingerprints of a mixture of three accessions per species were analysed by pooling 5 μL of the final selective
amplification products according to Mhameed et al. (1997). The low level of variation between individual samples showed that pooling accessions was justified. Three primer combinations (E +
ACCA/M + CATG, E + ACCT/M + CATC and E + AGCC/M + CACC) were selected from a test of 96 primer combinations, and these produced 272, 211 and 233 bands, respectively (Table 2). The choice of
the primers used in the study was based upon the visual clarity of banding patterns generated and a preferably low fingerprint complexity. The complexity of the banding pattern is a major
limiting factor for scoring AFLP fingerprints of large-size genomes. DATA ANALYSIS Positions of unequivocally visible and polymorphic AFLP markers were transformed into a binary matrix, with
‘1’ for the presence, and ‘0’ for the absence of a band at a particular position. The genetic distance (GD) between species was based on pair-wise comparisons and calculated according to
the equation: GD_xy_=1 − [2_N__xy_/(_N__x_ + _N__y_)], where _N__x_ and _N__y_ are the numbers of fragments to individuals _x_ and _y_, respectively, and _N__xy_ is the number of fragments
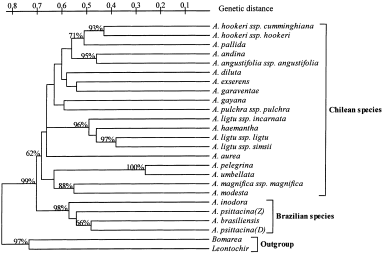
shared by both (Nei & Li, 1979). Genetic distances were computed by the software package TREECON (v. 1.3b) (Van De Peer & De Wachter, 1993). The dendrogram of the 22 _Alstroemeria_
species, the interspecific hybrid, _Bomarea_ and _Leontochir_ was generated based on the GD matrix by using cluster analysis, the UPGMA (unweighted pair group method using arithmetic
averages) method with 1000 bootstraps (Sneath & Sokal, 1973; Felsenstein, 1985) (Fig. 1). Principal co-ordinate analysis was performed to access interspecies relationships based on the
Nei & Li (1979) coefficient [2_N__xy_/(_N__x_ + _N__y_)] using the NTSYS-PC program (Rohlf, 1989). RESULTS AND DISCUSSION The average genetic distance among species excluding _Bomarea_,
_Leontochir_, the interspecific hybrid and _A. umbellata_ was 0.65 GD (a table showing the genetic distances between all the species studied is available from the authors on request).
_Alstroemeria umbellata_ was excluded because the accessions used were found to be highly related and possibly wrongly classified as different from _A. pelegrina_. The average GD among
accessions within a species was 0.32 GD (data not shown). In addition, the average GD between Brazilian species (GD: 0.27) and between Chilean species (GD: 0.33) was not significantly
different. Buitendijk & Ramanna (1996) suggested that the Chilean and Brazilian species form distinct lineages. The genetic diversification of _Alstroemeria_ species as detected by the
AFLP technique revealed three main clusters with 99% bootstrap values: the Chilean species, the Brazilian species and the outgroup (Fig. 1). This finding would support an early divergence of
these groups and is consistent with the occurrence of interspecific crossing barriers between the Chilean and Brazilian species (De Jeu & Jacobsen, 1995; Lu & Bridgen, 1997). The
variance of the first three principal co-ordinates accounted for 34.9% of the total variation, differentiated effectively among the species and reflected the main clustering of the
dendrogram. From the principal co-ordinate plot, four groups were clearly demarcated: (i) the Brazilian group; (ii) the Chilean group; (iii) the _A. ligtu_ group; and (iv) the outgroup (Fig.
2). The Brazilian species (_A. brasiliensis_, _A. psittacina_ and _A. inodora_) were consistently assigned to one cluster with 98% bootstrap values, whereas the Chilean species were rather
weakly clustered with 62% bootstrap values containing several subgroups within the Chilean group (Fig. 1 and 2). The dispersion of the Chilean species on the principal co-ordinate plot
reflected a wider genetic variation than the Brazilian species. However, the narrow variation of the Brazilian species might be caused by the limited number of species investigated.
Buitendijk & Ramanna (1996) described the similarities between C-banding patterns of _A. inodora_ and _A. psittacina_; in our study these species clustered strongly, reinforcing this
finding (Fig. 1). The similarity between _A. psittacina_ and _A. inodora_ was also revealed by allozyme analysis (Meerow & Tombolato, 1996) and by a study using species-specific
repetitive probes (De Jeu et al., 1995). These findings are also supported by the fact that _A. inodora_ and _A. psittacina_ are easily crossed (De Jeu & Jacobsen, 1995). In addition,
the Chilean species _A. aurea_ was positioned between three subgroups (Fig. 2). The unique position of _A. aurea_, and the observation that this species has a wide geographical spread,
suggest that other Chilean species may have evolved from _A. aurea_ ecotypes. _Alstroemeria aurea_ is indeed a widespread inhabitant in the regions with higher rainfall at the more southern
latitudes between 33 and 47°S in Chile (Bayer, 1987; Buitendijk & Ramanna, 1996). It is not found in Brazil, although _A. aurea_ plants are found on both sides of the Andes mountains in
Argentina, supporting the possibility that _A. aurea_ ecotypes were also the ancestors of the Brazilian species (A.F.C. Tombolato, personal communication). _Alstroemeria pelegrina_ and _A.
umbellata_ were assigned as sister species with a GD of 0.26 showing a remarkable genetic similarity (data available on request). The species we coded under the name _A. umbellata_ actually
seemed to be an _A. pelegrina_ species that did not flower for many years. _Alstroemeria haemantha_ was assigned to a group together with _A. ligtu_ ssp. _ligtu_, _A. ligtu_ ssp. _incarnata_
and _A. ligtu_ ssp. _simsii_ (Figs 1 and Fig. 2) (Aker & Healy, 1990; Ishikawa et al., 1997). Bayer (1987) suggested the synonymous name of _A. ligtu_ ssp. _ligtu_ for _A. haemantha_
Ruiz and Pavon. Our results support this hypothesis. _Alstroemeria exserens_ was positioned between the Chilean group and the _A. ligtu_ group (Fig. 2). _Alstroemeria andina_ and _A.
angustifolia_ ssp. _angustifolia_, and _A. hookeri_ ssp. _cumminghiana_ and _A. hookeri_ ssp. _hookeri_ were clustered together with 95% and 93% bootstrap values, respectively. The
interspecific hybrid (A1P2–2) was included in our study in order to investigate the possibility of the identification of the parental genotypes. The F1 hybrid A1P2–2 showed a 0.45-GD value
with _A. inodora_ and 0.59 GD value with _A. aurea_ showing genomic contribution of both parents (data available on request). It indicated the feasibility of the AFLP technique as a tool for
the identification of parental genotypes (Sharma et al., 1996; Marsan et al., 1998). _Bomarea_ and _Leontochir_ showed the mean GD value of 0.83 as the outgroup, thus showing large genetic
distances within the Alstroemeriaceae family. In conclusion, the genetic variation and the genetic relationships among _Alstroemeria_ species were efficiently rationalized by using AFLP
markers for the characterization of germplasm resources. In general, the topologies of the dendrogram and the principal co-ordinate analysis of our study were in agreement with Bayer’s views
(Bayer, 1987) on the classification of the _Alstroemeria_ species. Furthermore, this technique might be useful for the identification of parental genotypes in interspecific hybrids.
REFERENCES * Aker, S. and Healy, W. (1990). The phytogeography of the genus _Alstroemeria_. _Herbertia_ 46: 76–87. Google Scholar * Anastassopoulos, E. and Keil, M. (1996). Assessment of
natural and induced genetic variation in _Alstroemeria_ using random amplified polymorphic DNA (RAPD) markers. _Euphytica_ 90: 235–244. Article CAS Google Scholar * Bayer, E. (1987). Die
Gattung _Alstroemeria_ in Chile. _Mitt Bot Staatsamml München_, 24: 1–362. Google Scholar * Buitendijk, J. H. and Ramanna, M. S. (1996). Giemsa C-banded karyotypes of eight species of
_Alstroemeria_ L. and some of their hybrids. _Ann Bot_, 78: 449–457. Article Google Scholar * Buitendijk, J. H., Pinsonneaux, N. A. C., van Donk, M. S. and Lammeren, A. A. M. (1995).
Embryo rescue by half-ovule culture for the production of interspecific hybrids in Alstroemeria. _Sci Hortic_, 64: 65–75. Article Google Scholar * Dahlgren, R. M. T. and Clifford, H. T.
(1982). _Monocotyledons A Comparative Study_. Academic Press, London. Google Scholar * de Jeu, M. J. and Jacobsen, E. (1995). Early postfertilization ovule culture in _Alstroemeria_ L. and
barriers to interspecific hybridization. _Euphytica_ 86: 15–23. Article Google Scholar * de Jeu, M. J., Lasschuit, J., Chevalier, F. and Visser, R. G. F. (1995). Hybrid detection in
_Alstroemeria_ by use of species-specific repetitive probes. _Acta Hortic_, 420: 62–64. Article CAS Google Scholar * Dubouzet, J. G., Murata, N. and Shinoda, K. (1997). RAPD analysis of
genetic relationships among _Alstroemeria_ L. cultivars. _Sci Hortic_, 68: 181–189. Article CAS Google Scholar * Felsenstein, J. (1985). Confidence limits on phylogenies: an approach
using the bootstrap. _Evolution_ 39: 783–791. Article Google Scholar * Frei, O. M., Stuber, C. W. and Goodman, M. M. (1986). Use of allozymes as genetic markers for predicting performance
in maize single cross hybrids. _Crop Sci_, 26: 37–42. Article Google Scholar * Han, T. H., van Eck, H. J., de Jeu, M. J. and Jacobsen, E. (1999). Optimization of AFLP fingerprinting of
organisms with a large genome size: a study on _Alstroemeria_ spp. _Theor Appl Genet_, 98: 465–471. Article Google Scholar * Hill, M., Witsenboer, H., Zabeau, M., Vos, P., Kesseli, R. and
Michelmore, R. (1996). PCR-based fingerprinting using AFLPs as a tool for studying genetic relationships in _Lactuca_ spp. _Theor Appl Genet_, 93: 1202–1210. Article CAS Google Scholar *
Hutchinson, J. (1973). _The Families of Flowering Plants_. Clarendon Press, Oxford. Google Scholar * Ishikawa, T., Takayama, T., Ishizaka, H., Ishikawa, K. and Mii, M. (1997). Production of
interspecific hybrids between _Alstroemeria ligtu_ L. hybrid and _A. pelegrina_ L. var. _rosea_ by ovule culture. _Breed Sci_, 47: 15–20. Google Scholar * Kardolus, J. P., van Eck, H. J.
and van Den Berg, R. G. (1998). The potential of AFLPs in biosystematics: a first application in _Solanum_ taxonomy. _Pl Syst Evol_, 210: 87–103. Article Google Scholar * Lu, C. and
Bridgen, M. P. (1997). Chromosome doubling and fertility study of _Alstroemeria aurea_ × _A. caryophyllaea_. _Euphytica_ 94: 75–81. Article Google Scholar * Marsan, P. A., Castiglioni, P.,
Fusari, F., Kuiper, M. and Motto, M. (1998). Genetic diversity and its relationship to hybrid performance in maize as revealed by RFLP and AFLP markers. _Theor Appl Genet_, 96: 219–227.
Article CAS Google Scholar * Meerow, A. W. and Tombolato, A. F. C. (1996). The _Alstroemeria_ of Itatiaia. _Herbertia_. 51: 14–21. Google Scholar * Mhameed, S., Sharon, D., Kaufman, D.,
Lahav, E., Hillel, J., Degani, C. and Lavi, U. (1997). Genetic relationships within avocado (_Persea americana_ Mill.) cultivars and between _Persea_ species. _Theor Appl Genet_, 94:
279–286. Article Google Scholar * Nei, M. and Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. _Proc Natl Acad Sci USA_, 76:
5269–5273. Article CAS Google Scholar * Paul, S., Wachira, F. N., Powell, W. and Waugh, R. (1997). Diversity and genetic differentiation among populations of Indian and Kenyan tea
(_Camellia sinensis_ (L.) O. Kuntze) revealed by AFLP markers. _Theor Appl Genet_, 94: 255–263. Article CAS Google Scholar * Picton, D. D. and Hughes, H. G. (1997). Characterization of
_Alstroemeria_ species using Random Amplified Polymorphic DNA (RAPD) analysis. _HortScience_ 32: 482–482 Abstract: 323. Article Google Scholar * Rogers, S. O. and Bendich, A. J. (1988).
Extraction of DNA from plant tissues. _Plant Mol Biol Manual_, 6: 1–10. Google Scholar * Rohlf, F. J. (1989). _NTSYS-Pc Numerical Taxonomy and Multivariate Analysis System_, version 1.80.
Exeter Publications, New York, NY. Google Scholar * Schut, J. W., Qi, X. and Stam, P. (1997). Association between relationship measures based on AFLP markers, pedigree data and
morphological traits in barley. _Theor Appl Genet_, 95: 1161–1168. Article CAS Google Scholar * Sharma, S. K., Knox, M. R. and Ellis, T. H. (1996). AFLP analysis of the diversity and
phylogeny of _Lens_ and its comparison with RAPD analysis. _Theor Appl Genet_, 93: 751–758. Article CAS Google Scholar * Sneath, P. H. A. and Sokal, R. R. (1973). _Numerical Taxonomy_.
W.H. Freeman, San Francisco, CA. Google Scholar * Soltis, P. S., Soltis, D. E. and Doyle, J. J. (1992). _Molecular Systematics of Plants_. Chapman & Hall, New York, NY. Book Google
Scholar * Tohme, J., Gonzalez, D. O., Beebe, S. and Duque, M. C. (1996). AFLP analysis of gene pool of a wild bean core collection. _Crop Sci_, 36: 1375–1384. Article CAS Google Scholar
* Uphof, J. C. T. (1952). A review of the genus _Alstroemeria_. _Plant Life_. 8: 37–53. Google Scholar * van Depeer, Y. and de Wachter, R. (1993). Treecon: a software package for the
construction and drawing of evolutionary trees. _Comput Applic Biosci_. 9: 177–182. CAS Google Scholar * Vos, P., Hogers, R., Bleeker, M., Reijans, M., Vandelee, T., Hornes, M. _et al_
(1995). AFLP: a new technique for DNA fingerprinting. _Nucl Acids Res_, 23: 4407–4414. Article CAS Google Scholar * Wilkin, P. (1997). _Leontochir ovallei_ Alstroemeriaceae. _Curtis’s Bot
Magazine_. 14: 7–12. Article Google Scholar Download references ACKNOWLEDGEMENTS The authors would like to thank Anja G.J. Kuipers and Jaap B. Buntjer for critical reading of the
manuscript and for helpful comments. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Laboratory of Plant Breeding, The Graduate School of Experimental Plant Sciences, Wageningen University, PO
Box 386, AJ Wageningen, NL-6700, The Netherlands Tae-Ho Han, Marjo de Jeu, Herman van Eck & Evert Jacobsen Authors * Tae-Ho Han View author publications You can also search for this
author inPubMed Google Scholar * Marjo de Jeu View author publications You can also search for this author inPubMed Google Scholar * Herman van Eck View author publications You can also
search for this author inPubMed Google Scholar * Evert Jacobsen View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to
Tae-Ho Han. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Han, TH., de Jeu, M., van Eck, H. _et al._ Genetic diversity of Chilean and Brazilian
_Alstroemeria_ species assessed by AFLP analysis. _Heredity_ 84, 564–569 (2000). https://doi.org/10.1046/j.1365-2540.2000.00682.x Download citation * Received: 21 June 1999 * Accepted: 15
November 1999 * Published: 01 May 2000 * Issue Date: 01 May 2000 * DOI: https://doi.org/10.1046/j.1365-2540.2000.00682.x SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative KEYWORDS * Alstroemeriaceae * _Bomarea_ * classification * Inca lily * _Leontochir_ * Monocotyledonae
