Rapd of controlled crosses and clones from the field suggests that hybrids are rare in the salix alba–salix fragilis complex
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT The polyploid _Salix alba–Salix fragilis_ hybrid complex is rather difficult to study when using only morphological characters. Most of the features have a low diagnostic value for
unambiguously identifying the hybrids, introgression patterns and population structures, though morphological traits have proved to be useful in making a hybrid index. Morphology and
molecular variation from RAPDs were investigated in several case studies on willows from Belgium. A thorough screening of full-sib progenies of interspecific controlled crosses was made to
select homologous amplification products. The selected amplified products proved to be useful in a principal coordinate analysis for the estimation of variability of hybrid progenies. On the
basis of genetic similarities and ordination analysis, a method for the identification of clones in the field was established using presumed pure species and presumed introgressants. The
chosen reference clones were checked against additional European samples of putative pure species to ensure the reliability of the method beyond a regional scale. The RAPDs suggested that
both species have kept their gene pools well separated and that hybridization actually does not seem to be a dominating process. The observation that molecular markers do not always follow
the morphological traits or allozyme data is discussed. SIMILAR CONTENT BEING VIEWED BY OTHERS CONSTRUCTION OF A BREEDING PARENT POPULATION OF _POPULUS TOMENTOSA_ BASED ON SSR GENETIC
DISTANCE ANALYSIS Article Open access 29 October 2020 CONSTRUCTION AND GENETIC CHARACTERIZATION OF AN INTERSPECIFIC RASPBERRY HYBRIDS PANEL AIMING RESISTANCE TO LATE LEAF RUST AND ADAPTATION
TO TROPICAL REGIONS Article Open access 14 September 2023 GENE FLOW BETWEEN WILD TREES AND CULTIVATED VARIETIES SHAPES THE GENETIC STRUCTURE OF SWEET CHESTNUT (_CASTANEA SATIVA_ MILL.)
POPULATIONS Article Open access 02 September 2022 INTRODUCTION _Salix alba_ L. (white willow), _Salix fragilis_ L. (crack willow) and their hybrid _S._ × _rubens_ Schrank are closely related
polyploid species (2x=76) with a widely sympatric distribution throughout Europe. They are dioecious but may reproduce as well sexually as by vegetative propagation. The willows are typical
tree-forming pioneer species in alluvial plains and riparian zones. The delimitation between these polyploid taxa relies on relatively few diagnostic features in the morphology.
Consequently, large overlaps exist which make it difficult to identify samples from the field unambiguously. In many cases the two species coexist in mixed stands. Artificial hybridization
is possible, but the taxonomic identity as well as the identification of their metapopulations remains difficult. Different elements such as the lack of qualitative diagnostic characters,
the frequent occurrence of intermediate morphological forms and the successful interspecific controlled crosses, support the hypothesis that _S. alba_ and _S. fragilis_ may hybridize
frequently in nature. Hybrids and introgressed hybrids seem to dominate in the field when considering morphology (De Bondt, 1996). Few studies on allozyme variation in _Salix_ species have
been performed, but they have already shed some light on the genetic structure of diploid and polyploid willow populations. In _S. silicicola_ Raup. and _S. alaxensis_ (Anders.) Cov. (2x=38)
from North America, the differentiation between populations was low and the estimates of gene flow based on _G_ST values were rather moderate (Purdy & Bayer, 1995). This is consistent
with the reviewed data on trees with a dioecious breeding system and wind-dispersed seeds (Hamrick & Godt, 1989). Factors that promote high levels of genetic diversity within populations
in _Salix_ species include dioecy, high fecundity, wind-dispersed seeds and long-lived clonal growth. Characteristic for _Salix_ might be the allelic evenness of allozyme distribution
(Brunsfeld et al., 1991) as well as the low probability of genetic drift (Purdy & Bayer, 1995). Allozymes in full-sib controlled crosses of polyploid _S. alba_ and _S. fragilis_ from
Belgium showed Mendelian segregation for certain enzymes, whereas other genes had fixed heterozygosity. Most of the enzymes could not be used as good markers at the species level because
differences were mainly in allele frequencies (Triest et al., 1998a). Based on the inferred genetic control of these allozymes, a survey of 239 clones of the morphological _S. alba_–_S.
fragilis_ complex indicated that most of the allelic differentiation occurred at the local level of small tributaries, but that there is allelic evenness among the catchment areas of a river
system (Triest et al., 1998b). Tributaries which are 10–25 km in length were proposed as the most likely entities for further examination of putative hybridization and events of allelic
fixation. Yet, allozymes could give no evidence of extensive hybridization in the field between _S. alba_ and _S. fragilis_ (Triest et al., 1999). Because phylogenetic studies of closely
related taxa, or individuals within taxa, are often limited by the large number of individuals to be considered in the comparison (Stewart, 1993), a well-designed and conceptual approach has
to be worked out using suitable markers. Molecular markers that discriminate between closely related taxa can be useful for estimating the extent of hybridization and introgression among
species (Perron et al., 1995; Khasa & Dancik, 1996). Substantial amounts of DNA polymorphism can be detected by PCR using single decanucleotide primers (Williams et al., 1990). Although
the value of RAPDs can be questioned (e.g. Smith et al., 1994; Harris, 1995; van de Zande et al., 1995), the technique is more reliable when assaying by a considerable number of primers and
selecting a few repeatable and clearly interpretable RAPD fragments (Furman et al., 1997). Analysing the heritability of the RAPD fragments in the progeny of artificial crosses undoubtedly
will increase their value as molecular markers. Recently, AFLP have also proved to be useful in generating polymorphic DNA fragments for the identification of willows (Beismann et al.,
1997). The objective of this study was to estimate the extent of hybridization in the _S. alba–S. fragilis_ complex through the subsequent analysis of both morphology and RAPDs in three
complementary sets of samples: (i) artificially created hybrid progeny between presumed pure species in order to select appropriate RAPD markers; (ii) a morphological continuum of _S. alba_,
_S._ × _rubens_ and _S. fragilis_, representing a morphological hybrid index, in order to select appropriate reference clones of presumed pure species and eventually also the F1 hybrid and
introgressed hybrids; and (iii) representative European reference clones of _S. alba_ and _S. fragilis_ in order to test the validity of the chosen species-specific markers beyond a regional
scale. MATERIALS AND METHODS PLANT MATERIALS The willow buds or juvenile leaves originated from three complementary sets of sampling. Set no. 1: controlled crosses (Table 1) of parental _S.
alba_ and _S. fragilis_ clones originating from Belgium were first studied for morphological variation and RAPD. The clones were collected from the field in 1986 and 1990 and further
cultivated at the Institute for Forestry and Game Management (IBW, Ministry of Flemish Community). The artificial interspecific crosses were performed in 1990 and the progeny were further
cultivated at the IBW. The alphabetical codes of the parental types given at that time have been maintained for ease of comparison. Set no. 2: a representative morphological continuum of _S.
alba_, _S._ × _rubens_ and _S. fragilis_ was investigated for morphological variation and RAPD. Forty-six clones from the field (upstream tributaries of the river Schelde catchment) were
collected in 1982 and grown clonally at the IBW. The clones were previously identified as belonging to a putative series of pure species and their morphological intermediates. The codes used
for the hybrid index are: A=_S. alba_, AH=_S. alba_ hybrid or introgressant, R=_S._ × _rubens_, FH=_S. fragilis_ hybrid or introgressant, F=_S. fragilis_. Set no. 3: additional European
reference materials were the IBW clones of _S. alba_ 81.707 (cv. Bredevoort, The Netherlands), 81.708 (cv. Lievelde, The Netherlands), 81.709 (cv. Het goor, The Netherlands), 81.713 (I3.58,
Italy), 81.716 (Oberer Inn 1, Germany), 81.722 (Bulg 1/64, Bulgaria) and of _S. fragilis_ 95.002 and 95.003 (Sweden). These European clones were checked for their morphology and RAPDs
against previously selected Belgian reference clones. METHODS MORPHOLOGICAL DATA The identification of the individual trees and the analysis of morphological characteristics was carried out
on all samples used to set up the reference system. UPOV guidelines provided a basis for the selection of willow characteristics to be considered (De Bondt, 1996). Features were bud length,
leaf length, leaf width, ratio of leaf length/leaf width, petiole length, degree of serration at leaf margin, the shape of the buds and degree of pubescence on various organs (stem, upper
part of the twigs, lower half of the twigs, bud tip, upper side of the leaf and lower side of the leaf). The measurements were standardized into five classes before ordination analysis. DNA
EXTRACTION AND PCR PROCEDURE Buds or sprouting willow leaves were collected, frozen in liquid nitrogen and stored at −80°C. Total plant DNA for PCR was isolated according to a slightly
modified protocol from Murray & Thompson (1980). Previously frozen plant material (up to 3 g) was ground in a mortar containing liquid nitrogen. Twenty mL of DNA extraction buffer (100
mM Tris pH 8, 1.4 M NaCl, 20 mM EDTA, 2% CTAB, 1% PVPP, 1% β-mercaptoethanol) were added and the mixture subsequently incubated at 65°C for 20 min. After cooling to room temperature, 20 mL
of chloroform-isoamylalcohol (24:1, v/v) (CI) were added. The mixture was then centrifuged for 10 min at 3200 _G_. After recovery of the upper phase, 0.1 vol. 10% CTAB and 1 vol. CI were
added. A second centrifugation was carried out at 3200 _G_ for 10 min. 1.2 vol. precipitation buffer (100 mM Tris pH=8, 20 mM EDTA, 2% CTAB) was added to the recovered upper phase. After an
incubation of at least 20 min at room temperature, the nucleic acids were pelleted for 15 min at 8200 _G_. The pellet was dissolved in 5 mL 1 M NaCl and once fully dissolved, 10 μL of a
10-mg/mL RNase solution were added. This mixture was incubated at 37°C for 30 min. The DNA was precipitated by adding 500 μL 4 M LiCl and 15 mL 100% EtOH (−70°C, 30 min), followed by a
centrifugation at 12 800 _G_ for 10 min. The DNA pellet was washed in 70% EtOH. After a final centrifugation at 12 800 _G_ for 10 min, the DNA pellet was dried and subsequently dissolved in
500 μL TE. DNA presence was monitored by subjecting samples to 1% agarose gel electrophoresis in 1× TBE buffer (Sambrook et al., 1989) and by visual assessment of band intensities compared
with DNA standards. The exact DNA concentration and the purity was determined by spectrophotometry. A set of 14 decanucleotides of arbitrary sequence (obtained from Operon Technologies Inc.,
Alameda, CA.) were selected according to their reproducibility from the 40 primers that were initially employed, namely OPA01, OPA02, OPA04, OPA05, OPA07, OPA14, OPA20, OPO04, OPO11, OPO12,
OPO15, OPO16, OPO19, OPO20. The 25 μL reaction mixture contained approximately 25 ng template DNA, 5 pmol of a single decanucleotide, 100 μM each of dNTPs (Pharmacia) and 1 unit Taq
polymerase in the incubation buffer provided by the manufacturer of the enzyme (Boehringer Mannheim). The mixture was overlaid with two drops of mineral oil. Amplification was achieved in a
Thermojet thermocycler (Eurogentec) programmed as follows: 1 cycle of 2 min at 94°C; 40 cycles of 45 s at 92°C, 1 min at 36°C, 2 min at 72°C; 1 cycle of 5 min at 72°C. Amplification products
were subjected to electrophoresis on 1% agarose gels in 1× TBE and detected by ethidium bromide staining. Molecular weights were estimated using lambda DNA (Boehringer Mannheim) digested
with _Pst_I (Eurogentec). At least three repeat reactions were analysed for all primers on every individual. DATA ANALYSIS The reproducibility of the amplification products was improved by
altering the concentrations of Mg2+, Taq polymerase and the amount of template and primer DNA. In order to verify that the plant organs do not affect individual amplification products,
extractions were performed on buds (October–January), young leaves (March) and older leaves (September–October). No differences were found between the amplification products from the same
individual. Reliability of the amplification products was also tested using two different DNA thermal cyclers, using the same temperature profiles, obtaining nearly completely matching
amplified bands. Sometimes, the minor bands were difficult to interpret, but if such fragments were clearly visualized in one of the repeats, they were considered as present. Principal
coordinate analysis (PCOORDA) was carried out using the NTSYS-PC program (Rohlf, 1993). Banding patterns for all primers were compiled and introduced as a single rectangular data matrix
consisting of presence/absence data. The data set was then transformed into a similarity matrix using the simple matching (SM) coefficient of resemblance. RESULTS MORPHOLOGICAL VARIABILITY
From the analysis of the morphological characteristics of buds, leaves and twigs, no clearly separated groups of species or hybrids could be revealed in the progeny of controlled crosses,
the representative clones of the hybrid index and the European reference clones. The PCOORDA analysis indicated that there are numerous intermediate positions between the more extreme groups
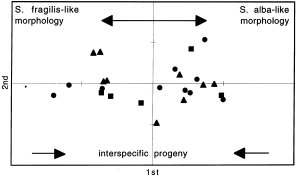
along the first principal coordinate. Therefore, only 27% and 7% of the morphological variation in the interspecific progeny could be explained by the first and second principal coordinate,
respectively (Fig. 1). When checking the position of the controlled crosses, it became clear that the progeny where the male _S. fragilis_ parent H was involved was morphologically very
variable. The interspecific A × H and B × H families had progenies with intermediate forms as well as with _S. alba_ morphology. The D × H family had progeny ranging morphologically from _S.
fragilis_ through intermediate forms to _S. alba_. The latter might indicate that the status of the parental type H is not a pure _S. fragilis_ but already an F1 or introgressed hybrid.
Therefore, D × H was considered as an inter- rather than an intraspecific cross for this analysis. In the representative clones of the hybrid index (Fig. 2), the _S. fragilis_ (F) and
so-called introgressed _S. fragilis_ hybrids (FH) could be identified rather consistently, whereas there could be more confusion among the _S. alba_ (A), _S._ × _rubens_ (R) and the
so-called introgressed _S. alba_ hybrids (AH). Only 21% and 9% of the variation could be explained by the first two axes. The previously chosen reference material of _S. fragilis_ (Sweden)
and _S. alba_ (The Netherlands, Germany, Italy and Bulgaria) were at opposite positions (Fig. 3) and confirmed the potential use of the morphological characters for identifying both species
and eventually their putative hybrids. As a result of the comparison with introgressed _S. alba_ clones, only 26% and 10% of the variation could be explained by the first two axes. RAPDS OF
CONTROLLED CROSSES (SET NO. 1) A total of 19 parental specific amplification products were selected out of 86 amplification products, using 13 primers (OPA01, OPA02, OPA04, OPA05, OPA07,
OPA14, OPA20, OPO04, OPO11, OPO12, OPO15, OPO16, OPO19). Estimation of the proportion of heterozygous RAPDs in the parental types through comparison of the presence/absence status for each
amplification product in their progeny revealed that _S. alba_ had more absence codes than _S. fragilis_ and consequently a lower proportion of observed heterozygosity (_H_o). The male
parental type H of _S. fragilis_ was heterozygous for all 10 of the markers used (Table 1). H thus could be close to an F1 in origin, explaining the large variability in the progeny. All of
the selected fragments were diagnostic of the individual parental types. Samples with a missing value were left out of the analysis presented here, because this might influence the position
of the intermediates in an ordination analysis (Fig. 4). All crosses resulted in progeny with a variable genotype composition. Only 20% and 16% of the variation could be explained by the
first and second principal coordinates, respectively. The most important observation for the further analysis of the hybrid _S. alba–S. fragilis_ complex is that the majority of the progeny
resulting from interspecific crosses between genotypically distinct parental types, were positioned in the intermediate zone and in between the parents. A minority of the progeny had
multi-RAPD genotypes similar to one of the parental types. RAPDS OF A REPRESENTATIVE MORPHOLOGICAL CONTINUUM (SET NO. 2) A total of 13 species-diagnostic amplification products were selected
from the 95 DNA fragments obtained with 12 primers (OPA02, OPA04, OPA05, OPA14, OPA20, OPO04, OPO11, OPO12, OPO15, OPO16, OPO19, OPO20). The reduction from 19 to 13 fragments was necessary
because some markers varied between individuals in the previous analysis of the controlled crosses. A PCOORDA analysis of 46 willow trees for these 13 amplification products using the simple
matching coefficient revealed that in this collection there were clearly two major groups corresponding to either _S. alba_ or _S. fragilis_ (Fig. 5). About 72% of the variation could be
explained by the first principal coordinate (only 8% for the second axis). As such, the morphological variability of 10 _S. alba_, 10 _S. alba_ hybrids, 10 _S._ × _rubens_, 10 _S. fragilis_
hybrids and six _S. fragilis_ was not reflected by these species-diagnostic genotypes. RAPDS IN REPRESENTATIVE EUROPEAN CLONES (SET NO. 3) The genetic similarity between individual clones of
diverse European origin and the selected Belgian reference clones was calculated using the same 13 diagnostic amplified products as in set no. 2. The similarity values were the lowest
between individuals of the putative pure _S. alba_ and the putative pure _S. fragilis_. Within a species, the lowest similarity values were 62% (for _S. alba_) and 46% (for _S. fragilis_).
Noteworthy is that the most deviating _S. fragilis_ (IBW no. 257) was more similar in these DNA fragments to _S. fragilis_ from Sweden (62%) than to all other samples. The Swedish willows,
however, are more similar to all other _S. fragilis_ samples than to IBW no. 257. The most deviating _S. alba_ (IBW no. 100) had its lowest similarity (15%) to the IBW no. 257 and to those
from Sweden (39%), but was more related to all other samples by over 50%. The PCOORDA revealed two clearly different clusters, each corresponding to _S. alba_ or _S. fragilis_ (Fig. 6).
About 77% of the variation on the first axis can be explained by the species specificity and positioned the reference clones as such, regardless of the origin. Only the clone from Bulgaria
was slightly differentiated from _S. alba_. The samples nos. 100 and 257, most differentiated from the _S. alba_ and _S. fragilis_ references, respectively, were placed towards intermediate
positions along the first principal coordinate but became separated along the second axis (explaining another 12% of the variation). They were included to frame the possible variability
(putative remnants of introgressed individuals) within each species. DISCUSSION The morphological analysis of the leaves, buds and twigs indicated that there are many potential hybrids in
the samples. This confirmed the observations of many botanists (Rechinger, 1964; De Langhe et al., 1983) that the majority of the willows of the _S. alba–S. fragilis_ complex are hybrids and
introgressants. Together with the ease of artificial cross-fertilization between both species this has even led to the suggestion that almost no pure species were available in the field as
a result of repeated introgression. Most of the quantitative morphological characteristics do not have a clear diagnostic value but are based on a continuous variation in the degree of
pubescence of the twigs, buds and leaves. Obviously, the identification of the least pubescent individuals (_S. fragilis_-like) can be achieved more consistently than of the more pubescent
_S._ × _rubens_ and _S. alba_. If the parental type H of _S. fragilis_ is an introgressant in origin, then the variation of its progeny can be better understood. Similarly, all other
individuals at an intermediate position of the ordination could be regarded as potential hybrids. The large proportion of morphological intermediates suggests that hybridization is a common
process between the species and that they coexist as a hybrid complex. When comparing the morphological variation of the progeny with RAPDs, using 19 diagnostic fragments at the level of the
individual parental types, it becomes clear that the more qualitatively chosen fragments in the parental types are providing sufficient information about the relationships between the
offspring and their parents. For the analysis of artificial hybrids, particular marker genes are better suited than morphology. In progeny studies, the availability of good markers is more
important than the total amount of fragments considered. The use of a few but unambiguous diagnostic bands gave clear information on marker segregation and estimations of heterozygosity in
these dominant markers. The large genetic variability of the progeny where H is involved as a male parent of _S. fragilis_ is consistent with the data on the morphological variability though
not on an individual basis. RAPDs were also reliable for showing that a large portion of the amplification products were heterozygous in most of the parental types. It thus can be expected
that extensive outcrossing between different genotypes should lead to a broad variability in the clones from the field. However, this broad variability seems not to be the case in the
present sampling or in a previous study (Triest et al., 1997). GENETIC DIFFERENTIATION VS. CONTINUUM IN MORPHOLOGY Using a morphological continuum of 46 clones, it was possible to trace
those trees that were at the largest genetic distance within the collection by means of comparing the original data from a similarity matrix and by using a PCOORDA. Much of the variation
could be explained in terms of a _S. alba_-like and _S. fragilis_-like grouping. Most samples were genetically close to the presumed references. In this collection of 46 trees, representing
a hybrid index, there were no samples observed with clear intermediate values of genetic similarities. A useful comparison of the morphological identity with the RAPDs of a previously
investigated series of the _S. alba–S. fragilis_ complex (Triest et al., 1997) showed that 41% of the individuals had a genotype and morphology both corresponding to _S. alba_ but that 23%
had a genotype typical of _S. alba_, but with a _S. fragilis_ morphology. Similarly, 28% of the individuals had a genotype and morphology corresponding to _S. fragilis_ but 8% had a genotype
typical of _S. fragilis_ and a morphology typical of _S. alba_. Because the progeny of controlled crosses were ordinated at positions between the parental types if these parents were
genetically divergent, then the putative F1 or introgressed hybrids (if they exist) from the field also should be positioned as intermediates. It might be questioned whether the comigrating
bands of individuals, originating from such different biogeographical regions, are really homologous. Nonhomology might introduce noise into the data set, eventually leading to inaccurate
estimates of genetic relationships. Rieseberg (1996) reviewed this problem and it can be shown that the chance for nonhomology is increased with larger genetic distances and when introducing
large randomly generated data sets. Because the selection of homologous amplification products was initially inferred from the controlled crosses of _Salix_, it can be argued that, in the
case of full-sibs, the comigrating fragments are indeed homologous for the samples from Belgium. Closely related individuals exhibiting a large (dominant) allelic evenness presumably were
less prone to RAPD artefacts. Allozymes in a similar set of _S. alba–S. fragilis_ clones from Belgium revealed a large allelic evenness without unique alleles in different regions (Triest et
al., 1998b). The additional willow trees from other European origins are clustered unambiguously among the presumed pure species of _S. alba_ and _S. fragilis_ of the Belgian clones. It is
evident that if the morphological identification of the Belgian clones was correct, the 13 diagnostic amplification products used here also had to be species-specific on a larger European
scale, which was demonstrated here. The observation that the above-mentioned European clones of putative pure _S. alba_ or _S. fragilis_ were genetically very close to the putative pure
Belgian clones might be indicative of a more widespread genetic evenness within both species, as well as of the genetic characteristics of the species. The latter are thus most likely
conservative over a large geographical area. When comparing the European references to the data obtained from the Belgian clones, previously used to set up a reference system (Triest et al.,
1997), it becomes evident that the genetic composition of the _S. alba–S. fragilis_ complex is disrupted, being skewed towards the pure species and not towards intermediate hybrid forms.
This dominating process, namely a low probability of hybrid formation and maintenance, could also explain why the species have kept their genetic identities quite well over such a large
geographical area. The possible role of introgression can be explained as an additional source of genetic variation. Hybridization and introgression increase the opportunities of
recombination by bringing together differentiated genomes. If these genomes are coadapted, recombination is unlikely to produce successful new variants, except perhaps in a disturbed or
changing environment. Many hybrid zones, however, appear to represent substantial barriers (in a so-called tension zone) to gene exchange, with limited introgression occurring in the face of
selection against hybrids or recombinants. Morphological observations do not always correspond with molecular analyses in the case of putative hybrids (Rieseberg & Brunsfeld, 1992;
Nason et al., 1992; Dawson et al., 1996). The utility of RAPDs in suggesting interspecific hybridization was clearly demonstrated in _Gliricidia_ where additional RFLPs revealed allelic
combinations consistent with a hybrid origin at similar localities (Dawson et al., 1996). The Poptun individuals of _Gliricidia_ were heterozygous at two RFLP loci and strongly indicated F1
hybrids. The observation that the suggested F1 hybrids were positioned as intermediates along the first principal coordinate of the PCOORDA, but grouped separately along the second principal
coordinate is quite similar to our observations on willows. Morphology and isozymes indicated a close relationship between _Solidago albopilosa_ and _S. flexicaulis_, whereas RAPDs on F1
hybrids indicated that _S. flexicaulis_ only hybridizes with _S. caesia_ (Esselman & Crawford, 1997). Phylogenetic relationships in 19 species of _Rosa_ apparently showed good
correlation with previous classifications based on morphological and karyological studies (Millan et al., 1996). Similarly, allozymes do not always follow morphological variation. In
_Aesculus_ (De Pamphilis & Wyatt, 1989) and _Populus_ (Keim et al., 1989) hybridization could be detected on the basis of diagnostic features. In _Quercus rubra_ and _Q. ellipsoides_,
morphological hybrids were observed but the isozyme polymorphism was similar in both species (Hokanson et al., 1993). In an outcrossing perennial (_Ipomopsis_), putative hybridization was
noted in the morphology, but again, the allozyme composition remained very similar among three species of a complex (Wolf et al., 1991). In some cases, molecular markers reveal hybrid forms
that were not expected on morphological grounds. The detection of hybrids and introgressed individuals between black spruce and red spruce was more efficient using molecular (RAPD) markers
than with morphological traits (Perron & Bousquet, 1997). Their morphological discriminant model, based on twig characters, misclassified as parental types many of the hybrid or
introgressant trees detected with molecular markers. They observed a higher proportion of hybrids with RAPDs than with morphology, which led them to state that natural hybridization is a
rule rather than an exception. In contrast, our results on the willows demonstrate that morphological traits indicate a higher level of hybrid formation than do the RAPD markers. This might
be explained by the mixed reproductive strategy in willows, that is in contrast to the highly sexually reproducing spruces. Perhaps chloroplast DNA markers are more promising for detecting
interspecific gene flow, as shown for American white oaks (Whittemore & Schaal, 1991). Both species of willows studied here may have populations that are structured largely
independently. It might be expected that _S. fragilis_ is more clonal than _S. alba_. If so, multiple local contacts would provide the only opportunity for many hybridization events. Such
mosaic hybrid zones are logical places to look for intermediate variants. Beismann et al. (1997) most likely observed such a local contact zone between _S. alba_ and _S. fragilis_ with
hybridization as a result. Clonality, however, must have influenced the genetic structuring of the mixed population, because they collected _S. fragilis_ mainly at the higher altitudes and
_S. alba_ at the lower altitudes. Surprisingly, the presumed hybrids they detected with AFLP markers seemed to be rather homogeneous, suggesting that they were F1 hybrids rather than
introgressed ones. Local mosaic patterns could be tension zones of coadapted gene complexes, although hybrid zones often have been reported to remain narrow, with a low frequency of
introgressive hybrids (Harrison, 1990). The present study could indicate that identification of morphological hybrids is not necessarily following the chosen species-specific RAPD
amplification products. Hybridization in the _S. alba–S. fragilis_ complex is suggested to be a process with a low occurrence in the lowlands of Belgium. REFERENCES * Beismann, H., Barker,
J. H. A., Karp, A. and Speck, T. (1997). AFLP analysis sheds light on distribution of two _Salix_ species and their hybrid along a natural gradient. _Mol Ecol_, 6: 989–993. Article CAS
Google Scholar * Brunsfeld, S. J., Soltis, D. E. and Soltis, P. S. (1991). Patterns of genetic variation in _Salix_ section _Longifoliae_ (Salicaceae). _Am J Bot_, 78: 855–869. Article
Google Scholar * Dawson, I. K., Simons, A. J., Waugh, R. and Powell, W. (1996). Detection and pattern of interspecific hybridization between _Gliricidia sepium_ and _G. maculata_ in
Meso-America revealed by PCR-based assays. _Mol Ecol_, 5: 89–98. Article CAS Google Scholar * De Bondt, R. (1996). _Role of Genetic Diversity and Identity in Replantations — Case Study
on_ Salix _sp_. MSc Thesis, Vrije Universiteit, Brussel. * De Langhe, J., Delvosalle, L., Duvigneaud, J., Lambinon, J. and Vanden Berghen, C. (1983). _Flora Van België, Het Groothertogdom
Luxemburg, Noord-Frankrijk en de aangrenzende Gebieden_ Nationale Plantentuin, Meise. * De Pamphilis, C. and Wyatt, R. (1989). Hybridization and introgression in Buckeyes (_Aesculus_:
Hippocastanaceae): a review of the evidence and a hypothesis to explain long-distance gene flow. _Syst Bot_, 14: 593–611. Article Google Scholar * Esselman, E. J. and Crawford, D. J.
(1997). Molecular and morphological evidence for the origin of _Solidago albopilosa_ (Asteraceae), a rare endemic of Kentucky. _Syst Bot_, 22: 245–257. Article Google Scholar * Furman, B.
J., Grattapaglia, D., Dvorak, W. S. and O’malley, D. M. (1997). Analysis of genetic relationships of central American and Mexican pines using RAPD markers that distinguish species. _Mol
Ecol_, 6: 321–331. Article CAS Google Scholar * Hamrick, J. L. and Godt, M. J. (1989). Allozyme diversity in plant species. In: Brown, A. H. D., Clegg, M. T., Kahler, A. L. and Weir, B.
S. (eds) _Plant Population Genetics, Breeding and Genetic Resources_, pp. 43–63. Sinauer, Sunderland, MA. Google Scholar * Harris, S. A. (1995). Systematics and randomly amplified
polymorphic DNA in the genus _Leucaena_ (_Leguminosae, Mimosoideae_). _Pl Syst Evol_, 197: 195–208. Article CAS Google Scholar * Harrison, R. G. (1990). Hybrid zones: windows on
evolutionary process. In: Futuyma, D. and Antonovics, J. (eds) _Oxford Surveys in Evolutionary Biology_, 7, 69–128, Oxford University Press. Google Scholar * Hokanson, S. C., Isebrands, J.
G., Jensen, R. J. and Hancock, J. F. (1993). Isozyme variation in oaks of the Apostle Islands in Wisconsin: genetic structure and levels of inbreeding in _Quercus rubra_ and _Q.
ellipsoidalis_ (Fagaceae). _Am J Bot_, 80: 1349–1357. Article CAS Google Scholar * Keim, P., Paige, K. N., Whitham, T. G. and Lark, K. G. (1989). Genetic analysis of an interspecific
hybrid swarm of _Populus_: occurrence of unidirectional introgression. _Genetics_ 123: 557–565. CAS PubMed PubMed Central Google Scholar * Khasa, P. D. and Dancik, B. P. (1996). Rapid
identification of white Engelmann spruce species by RAPD markers. _Theor Appl Genet_, 92: 46–52. Article CAS Google Scholar * Millan, T., Osuna, F., Cobos, S., Torres, A. M. and Cubero,
J. I. (1996). Using RAPDs to study phylogenetic relationships in _Rosa_. _Theor Appl Genet_, 92: 273–277. Article CAS Google Scholar * Murray, M. and Thompson, W. (1980). Rapid isolation
of high molecular weight plant DNA. _Nucl Acids Res_, 8: 4321–4321. Article CAS Google Scholar * Nason, J. D., Ellstrand, N. C. and Arnold, M. L. (1992). Patterns of hybridization and
introgression in populations of oaks, manzanitas and irises. _Am J Bot_, 79: 101–111. Article Google Scholar * Perron, M. and Bousquet, J. (1997). Natural hybridization between black
spruce and red spruce. _Mol Ecol_, 6: 725–734. Article Google Scholar * Perron, M., Gordon, A. and Bousquet, J. (1995). Species-specific RAPD fingerprints for the closely related _Picea
maritima_ and _P. rubens_. _Theor Appl Genet_, 91: 142–149. Article CAS Google Scholar * Purdy, B. G. and Bayer, R. J. (1995). Allozyme variation in the Athabasca sand dune endemic,
_Salix silicola_ and the closely related widespread species, _S. alaxensis_. _Syst Bot_, 20: 179–190. Article Google Scholar * Rechinger, K. H. (1964). _Salicaceae_. In: Tutin, T. G.,
Heywood, V. H., Burges, N. A., Valentine, D. H., Walters, S. M. and Webb, D. A. (eds) _Flora Europaea_, vol. 1, pp. 43–55. Cambridge University Press, Cambridge. Google Scholar * Rieseberg,
L. H. (1996). Homology among RAPD fragments in interspecific comparisons. _Mol Ecol_, 5: 99–105. Article CAS Google Scholar * Rieseberg, L. H. and Brunsfeld, S. J. (1992). Molecular
evidence and plant introgression. In: Soltis, D. E., Soltis, P. S. and Doyle, J. J. (eds) _Plant Molecular Systematics_, pp. 151–176. Chapman & Hall, New York. Chapter Google Scholar *
Rohlf, F. J. (1993). Ntsys-Pc. _Numerical taxonomy and multivariate analysis system version 1.80_. Exeter Software, Applied Biostatistics Inc., New York. Google Scholar * Sambrook, J.,
Fritsch, E. F. and Maniatis, T. (1989). _Molecular Cloning: A Laboratory Manual_, 2nd edn. Cold Spring Harbor Laboratory Press, NY. Google Scholar * Smith, J. J., Scott-Craig, J. S.,
Leadbetter, J. R., Bush, G., Roberts, D. L. and Fullbright, D. W. (1994). Characterization of random amplified polymorphic DNA (RAPD) products from _Xanthomonas campestris_ and some comments
on the use of RAPD products in phylogenetic analysis. _Mol Phylogenet Evol_, 3: 135–145. Article CAS Google Scholar * Stewart, C. (1993). The powers and pitfalls of parsimony. _Nature_
361: 603–607. Article CAS Google Scholar * Triest, L., de Greef, B., de Bondt, R., Vanden Bossche, D., D’haeseleer, M., van Slycken, J. _et al_ (1997). Use of RAPD markers to estimate
hybridization in _Salix alba_ and _Salix fragilis_. _Belg J Bot_, 129: 140–148. Google Scholar * Triest, L., de Greef, B., D’haeseleer, M., Echchgadda, G., van Slycken, J. and Coart, E.
(1998a). Variation and inheritance of isozyme loci in controlled crosses of _Salix alba_ and _Salix fragilis_. _Silvae Genet_, 47: 88–94. Google Scholar * Triest, L., de Greef, B., van
Slycken, J. and Coart, E. (1998b). Isozyme differentiation between sympatric clones of _Salix alba_ and _Salix fragilis_. _For Genet_ 5: 249–260. Google Scholar * Triest, L., de Greef, B.,
Vermeersch, S., van Slycken, J. and Coart, E. (1999). Genetic variation and putative hybridisation in _Salix alba_ and _Salix fragilis_: Evidence from allozyme data. _Pl Syst Evol_, 215:
169–187. Article Google Scholar * Van de Zande, L., Bijlsma, R. and van de Zande, L. (1995). Limitations of the RAPD technique in phylogeny reconstruction in _Drosophila_. _J Evol Biol_,
8: 645–656. Article CAS Google Scholar * Whittemore, A. T. and Schaal, B. A. (1991). Interspecific gene flow in oaks. _Proc Natl Acad Sci USA_, 88: 2540–2544. Article CAS Google Scholar
* Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A. and Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. _Nucl Acid
Res_, 18: 6531–6535. Article CAS Google Scholar * Wolf, P. G., Soltis, P. S. and Soltis, D. E. (1991). Genetic relationship and patterns of allozyme divergence in the _Ipomopsis
aggregata_ complex and related species (Polemoniaceae). _Am J Bot_, 78: 515–526. Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This project was funded by the Fund for
Scientific Research, Flanders (mandate of Senior Research Associate and contract no. S2/5-ID.E 53 on ecogenetic diversity in plant populations), the Ministry of the Flemish Community (Dienst
Bos en Groen, contract nos WB/10/94 and BG/17/95 on genetic diversity in tree species) and the Vrije Universiteit Brussel (OZR funding). Special thanks to Dirk Vanden Bossche for technical
assistance and to Els Coart (Institute for Forestry and Game Management, Ministry of the Flemish Community) for providing the controlled crosses and the reference clones. AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Laboratorium voor Algemene Plantkunde en Natuurbeheer (General Botany and Nature Management), Vrije Universiteit Brussel, Pleinlaan 2, Brussels, B-1050, Belgium
Ludwig Triest, Bart de Greef & Ruth de Bondt * Jos Van Slycken, Institute for Forestry and Game Management, Ministry of the Flemish community, Gaverstraat 4, Geraardsbergen, 9400,
Belgium Jos van Slycken Authors * Ludwig Triest View author publications You can also search for this author inPubMed Google Scholar * Bart de Greef View author publications You can also
search for this author inPubMed Google Scholar * Ruth de Bondt View author publications You can also search for this author inPubMed Google Scholar * Jos van Slycken View author publications
You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Ludwig Triest. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE
THIS ARTICLE Triest, L., de Greef, B., de Bondt, R. _et al._ RAPD of controlled crosses and clones from the field suggests that hybrids are rare in the _Salix alba–Salix fragilis_ complex.
_Heredity_ 84, 555–563 (2000). https://doi.org/10.1046/j.1365-2540.2000.00712.x Download citation * Received: 23 June 1999 * Accepted: 09 November 1999 * Published: 01 May 2000 * Issue Date:
01 May 2000 * DOI: https://doi.org/10.1046/j.1365-2540.2000.00712.x SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry,
a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * genetic diversity *
hybridizatiom * polymorphism * RAPD * _Salix alba_ * _Salix fragilis_
