Apc gene hypermethylation and prostate cancer: a systematic review and meta-analysis
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Prostate cancer (PCa) is a worldwide disease that affects a large number of males. Although prostate-specific antigen (PSA) screening is used, the specificity is limited. This study
analyzes the sensitivity and specificity of adenomatous polyposis coli (APC) methylation for PCa detection in body fluids and tissues. Combining search results from PubMed and Embase, 19
studies were included, 5 involving body fluids and 14 involving prostate tissue, with 2344 subjects. In body fluid subgroups, the pooled sensitivity and specificity was 0.53 (95% confidence
interval (CI): 0.28–0.78) and 0.92 (95% CI: 0.86–0.95), respectively. From tissue studies, the results presented as 0.84 (95% CI: 0.70–0.92) and 0.91 (95% CI: 0.77–0.97). To confirm the
results, we conducted a further analysis by removing studies which introduced high heterogeneity due to the type of cases and controls. The same degree of sensitivity and specificity was
presented in two subgroups (urine: sensitivity 0.46, 95% CI: 0.39–0.53; specificity 0.87, 95% CI: 0.64–0.96; tissue: sensitivity 0.87, 95% CI: 0.72–0.94; specificity 0.89, 95% CI:
0.68–0.97). In addition, analysis of the interaction between APC methylation and PCa showed strong association in the whole data set (odds ratio (OR)=24.91, 95% CI: 12.86–48.24, _I_2=72.5%).
Pooling the same two main subgroups (tissue/fluid) gave a pooled OR of 33.54 (95% CI: 14.88–75.59; _I_2=70.7%) and 8.20 (95% CI: 2.84–23.74, _I_2=64.2%), respectively. From this study, the
results suggest that APC promoter methylation may be the potential testing for PCa diagnosis and provide a new viewpoint in the treatment of PCa. SIMILAR CONTENT BEING VIEWED BY OTHERS A
GENETIC STUDY TO IDENTIFY PATHOGENIC MECHANISMS AND DRUG TARGETS FOR BENIGN PROSTATIC HYPERPLASIA: A MULTI-OMICS MENDELIAN RANDOMIZATION STUDY Article Open access 04 October 2024 URINE
BIOMARKERS CAN PREDICT PROSTATE CANCER AND PI-RADS SCORE PRIOR TO BIOPSY Article Open access 05 August 2024 DETECTION OF COLORECTAL CANCER IN URINE USING DNA METHYLATION ANALYSIS Article
Open access 27 January 2021 INTRODUCTION Prostate cancer (PCa) is one of the most common cancers in the western world1 and is said to be the most frequently detected male cancer and the
second most frequent cause of male cancer deaths.2 In 2009, it was suggested that one in six men would be affected, involving 192 280 new cases of PCa and 27 360 PCa-related deaths in the
United States.3 However, significant symptoms can be found in only about half of all diagnosed patients, which indicate the low diagnosis and high mortality rate.4 With an increasing rate of
morbidity of 3% per year over several decades,5 reliable methods of diagnosis are needed urgently. In the 1990s, prostate-specific antigen (PSA) testing became widespread,6 which provided a
new approach in the diagnosis of PCa. Disappointingly, although serum PSA is generally used in PCa screening, the poor baseline values and low specificity limits its functions.7 DNA
methylation of gene promoters may provide the ideal works. There are important advantages of using DNA methylation as cancer biomarkers. In particular, methylated DNA can be detected with a
high degree of specificity and sensitivity,8 which promotes its application to minimal samples from PCa patients. To date, over 50 hypermethylated loci have been identified in PCa.9, 10
Among these loci, adenomatous polyposis coli (APC) is a well-characterized tumor-suppressor gene. The _APC_ gene is located on the long (q) arm of chromosome 5 between positions 21 and 22,
between 112 118 468 and 112 209 532 base pairs (bp). Methylation of the genes is associated with PCa. The purpose of this study was to conduct a meta-analysis of the sensitivity and
specificity of APC methylation on PCa detection in body fluid (blood and urine) and prostate tissues. The results of this article will help to provide a reliable biomarker for the diagnosis
and discrimination of PCa. We also determined whether APC methylation was correlated with pathological stage, Gleason score and PSA level among the cases. MATERIALS AND METHODS STUDY
SELECTION We conducted a comprehensive literature search of PubMed and Embase databases using the keywords ‘prostate cancer’, ‘PCa’, ‘prostate adenocarcinoma’, ‘APC’ and ‘adenomatous
polyposis coli’. Additional studies were found via the reference lists of the identified articles. The last retrieval was conducted in October 2012. Our inclusion criteria were as follows:
(1) measurement of DNA methylation in one of the following samples: blood, plasma, serum, buffy coat, urine, ejaculates, or prostate tissues; (2) a case–control study; and (3) published in
English language. Our exclusion criteria were: (1) APC methylation conducted in the cell lines; (2) unavailable raw data on the amount of methylation among cases and controls, respectively
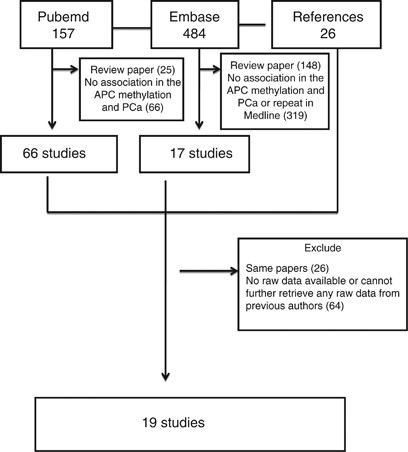
(some studies reported specificity and sensitivity without the exact counts); (3) review paper. The selection process for studies included in our review is shown in Figure 1. Our search
strategy and application of the inclusion/exclusion criteria resulted in a total of 19 articles that were included in the systematic review.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29 A description of the included studies is given in Tables 1 and 2. The following data were recorded for each study: author’s name, year of publication, sample
forms, method, 5′-3′ primers (forward and reverse, respectively), amplicon size (bp) and annealing temperature (°C), country, race, cancer clinical classification, PSA, Gleason score, type
of cases and controls, type of PCR method and other relevant characteristics of the study population. SENSITIVITY AND SPECIFICITY ANALYSIS The normalized index of methylation (NIM) and
receiver operator characteristic (ROC) curves were applied in most analyses. NIM was a color-scaled figure in which white represented NIM of zero (no methylation detected) and red defined a
NIM of 0.99 (99% of input DNA is methylated). NIM was defined in any given sample to be the ratio of the normalized amount of methylated templates at the promoter of interest to the
normalized amount of converted MYOD1 templates (NIM=[(GENE sample)/(GENE SssI)]/[(MYODI sample)/(MYOD SssI)]). Here, GENE sample and GENE SssI were said to be the number of entirely
methylated copies of the gene of interest in a given sample. Similar definitions were applied to MYOD1 sample and MYOD1 SssI. In addition, the optimal threshold for methylation was
determined on the basis of the area under the ROC curve.26 Combining the NIM and ROC curve; the numbers of the methylation were recorded. Before we conducted sensitivity and specificity
analyses, the amount of case and control methylation were collected. Among the included studies, two categories were assigned as controls: (1) patients who had negative biopsies but had
other diseases including benign prostatic hyperplasia (BPH), and (2) healthy controls. Nevertheless, biopsy-confirmed PCa and high-grade prostatic intraepithelial neoplasia (HGPIN) were
treated as cases. Thus, the true-positive (TP) samples were limited to those that had methylation within the exact cases. Meanwhile, in the case samples, the false-positive (FP) ones were
indicated to have no methylation. The same definitions was given for true negative (TN) and false negative (FN) in controls. All analyses were conducted with the Midas system in Stata. Owing
to the different types of samples in the eligible studies, we conducted a further analysis to present more robust results on APC methylation as the detection marker. In this analysis, the
fluid and tissue subgroups were processed. The types of the samples were described in detail. Among the tissue subgroup, we excluded studies that had other cancer diseases samples (such as
lung cancer and bladder cancer) because APC methylation might be also expressed in the different cancers. The analysis of the sensitivity and specificity was then conducted as above. The
same processes were applied in the fluid subgroup. ASSOCIATION ANALYSIS Odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were used to describe the effect of the
association between APC methylation and PCa, pathological stage, Gleason score, and PSA levels. The pathological stages were categorized into two subgroups: T1/T2 and T3/T4.27 For the
Gleason score, a score of 7 was used as a cutoff. PSA levels were dichotomized as less or greater than 4 ng/ml. On the basis of individual study ORs, pooled OR was estimated. According to
the heterogeneity statistic _I_2, a fixed effect or a random-effects model was selected: a fixed effect model was used when _I_2<50%, otherwise a random-effects model was used. In
addition, when the results of the constituent studies differed among themselves, the effects incorporated an estimate of the inter-study variance and therefore provided wider 95% CI. At the
same time, the _I_2-based _Q_ statistic was used, which describes the weighted sum of the squared difference between the overall effect size and the effect size from each study, to assess
heterogeneity (_P_<0.10 as the standard).30 ROC ANALYSIS To assess whether variation in the threshold definition of a positive result produced an association between sensitivity and
specificity values across studies, we calculated the summary receiving operating characteristic (S-ROC) curve.31 The logits of the TP and FP rates were used to estimate the linear regression
of the log-OR from each study. Independent analysis of pooled sensitivity and specificity using standard methods for binary data were used when the regression between these quantities was
null. All data used a log-odds scale (eg, for specificities, the effect size used was log (Spec/(1-Spec)).32 Finally, due to the heterogeneity between studies, we performed the Cochrane Q
test of heterogeneity for each analysis (based on deviations of observed log-odds from the common log-odds). The analyses were conducted using Stata 9.0 (Stata Corporation, College Station,
TX, USA), and all _P_-values were two-tailed. RESULTS CHARACTERISTICS After retrieving search results from the PubMed and Embase databases with the associated keywords, there were 157 and
484 articles retrieved from PubMed and Embase, respectively, on PCa and APC methylation/gene. Among these, we identified 95 relevant studies that described PCa and APC methylation. There
were 11 and 29 articles that used fluid (urine, blood or others) samples and tissues, respectively; 55 references were conference abstracts, non-experimental studies or otherwise not
available, and were removed, resulting in 40 articles. While reading the full texts, 21 articles were removed. Six could not provide data on the methylation among fluid samples (urine and
blood), and33, 34, 35, 36, 37, 38 15 of the 29 articles using tissues had no raw data (the specific numbers of cases and controls with methylation were unclear, which made it difficult to
calculate the data required for sensibility and specificity) or studied other factors such as the _TMPRSS2_ gene and urothelial carcinomas.39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51,
52, 53 Finally, 19 studies met the inclusion criteria and were included.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Among these 19 studies, 5 involved body
fluid (blood, urine and so on)11, 12, 13, 14, 15 and the remaining 14 articles involved sample tissues.16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Ten studies used quantitative
real-time methylation-specific PCR (QMSP) to detect APC methylation,11, 13, 14, 18, 19, 21, 22, 23, 25, 26 seven were conducted using the method of methylation-specific PCR (MSP),12, 15, 17,
20, 24, 27, 29 and two studies used pyrosequencing.16, 28 All cases were of PCa or HGPIN and were hospitalized, while controls were limited to be BPH, healthy subjects or those who had
genitourinary cancer (bladder carcinoma) with a healthy prostate. In regard to the type of cases, 14 studies were PCa,11, 12, 13, 14, 15, 16, 18, 19, 20, 23, 24, 27, 28, 29 and five were a
mixture of PCa, HGPIN and metastasis or other cancer tissues.17, 21, 22, 25, 26 Among the controls, eight studies were normal, biopsy negative, or non-tumor12, 13, 14, 15, 17, 25, 26, 29
nine were BPH,16, 18, 19, 20, 21, 23, 24, 27, 28 and two were a combination of BPH, normal subjects and bladder carcinoma.11, 22 In addition, 14 were Caucasian,11, 12, 13, 14, 15, 18, 19,
20, 21, 22, 23, 25, 26, 29 four were Asian,16, 17, 24, 27 and one involved mixed races from different continents28 who came from United States, United Kingdom, Portugal, Germany, Korea,
Japan, France, and China. SPECIFICITY AND SENSITIVITY OF APC PROMOTER METHYLATION USING DIFFERENT TYPES OF SAMPLES All the results are shown in Table 3. The pooled specificity for all
included studies was 0.91 (95% CI: 0.82–0.95), and the pooled sensitivity was 0.78 (95% CI: 0.63–0.88). For the traditional biomarker, the sensitivity of PSA varied, but the specificity was
generally low at about 20%,32, 54 which suggested that the APC methylation test has a much higher specificity than the PSA test. There was no evidence of publication bias (_P_=0.33). In
addition, we classified all samples into two groups according to specimen type (fluid/tissue). Among the fluid studies (urine/blood), the pooled sensitivity and specificity was 0.53 (95% CI:
0.28–0.78) and 0.92 (95% CI: 0.86–0.95), respectively. For the tissue studies, the pooled sensitivity and specificity was 0.84 (95% CI: 0.70–0.92) and 0.91 (95% CI: 0.77–0.97),
respectively. Publication bias results are showed in Table 3. The S-ROC curve is showed in Figure 2. In the extra analysis of the sensitivity and specificity, among the studies, we excluded
those that would likely introduce high heterogeneity due to different types of cases and controls. Finally, one study13 and four articles19, 20, 22, 26 in the fluid and tissue subgroups,
respectively, with different samples. The pooled specificity (0.90, 95% CI: 0.77–0.96) and pooled sensitivity (0.78, 95% CI: 0.59–0.90) were similar to the previous results. In addition, in
the urine subgroup, the sensitivity was lower (0.45, 95% CI: 0.39–0.53) and the specificity was 0.92 (95% CI: 0.84–0.96). The similar results were presented in the tissue subgroups
(sensitivity 0.87, 95% CI: 0.72–0.94; specificity 0.89, 95% CI: 0.68–0.97), which suggested a high level of sensitivity and specificity. ASSOCIATION BETWEEN APC PROMOTER METHYLATION AND
PATHOLOGICAL STAGE, GLEASON SCORE, AND PSA LEVELS IN PCA CASES We also conducted an analysis of the relationship between the pathological stage, Gleason score, and PSA levels among PCa cases
and APC promoter methylation. Details are shown in Table 4. We found no significant association between groups with the appropriate models except for the pathological stage (OR=0.42, 95%
CI: 0.25–0.70, _I__2_=0.0%). Finally, the association between APC promoter methylation and PCa was conducted and the pooled OR was 24.91 (95% CI: 12.86–48.24, _I_2=72.5%), with pooled ORs of
33.54 (95% CI: 14.88–75.59, _I_2=70.7%) and 8.20 (95% CI: 2.84–23.74, _I_2=64.2%) in the tissue and fluid groups, respectively (Figure 3). DISCUSSION DESCRIPTION Although there were have
been many studies about of the sensitivity and specificity of APC promoter methylation in relation to PCa, a summary meta-analysis has not been reported. To confirm the real function of APC
promoter methylation in predicting PCa, this study is required. This meta-analysis is based on 19 studies containing a total of 1600 cases and 744 controls. The major finding of this study
has demonstrated that APC promoter methylation may be associated with PCa. With the high sensitivity and specificity, it would be an ideal biomarker in diagnosing PCa. POOLED SPECIFICITY AND
SENSITIVITY ANALYSIS With the strong association (OR=24.91, 95% CI: 12.86−48.24, _I_2=72.5%), we conducted the specificity and sensitivity analysis. For all included studies, either in the
whole data set (pooled specificity 0.90, 95% CI: 0.80–0.95; pooled sensitivity 0.78, 95% CI: 0.63–0.88) or from subgroup analysis (fluid: specificity 0.92, 95% CI: 0.86–0.95; sensitivity
0.53, 95% CI: 0.28–0.78; tissue: specificity 0.91, 95% CI: 0.77–0.97; sensitivity 0.84, 95% CI: 0.70–0.92), the specificity and sensitivity of the test seemed to be high as an biomarker,
which suggests a greater directive function in diagnosis with the biomarker of APC methylation. However, due to differences between cases and controls between studies, we removed some
studies to make the data more homogeneous. These results suggested no major changes, and therefore provide support to our conclusion. As we have proposed above, the PSA test had varying high
sensitivity and poor specificity (about 20%). Therefore, according to our results, APC methylation with high specificity may increase the veracity of diagnosis when combined with the PSA
test. For sensitivity, only the fluid subgroup was lower than the PSA test. Due to the high pooled specificity, we propose the following: (1) because of the high sensitivity of the PSA test,
potential patients would be screened out. Likewise, APC promoter methylation would need to be detected with high specificity among those patients who were positive in the PSA test. If the
results of both tests were elevated among patients, future biopsies may be warranted. In this way, not only would the diagnosis be elevated, but also the weakness of the PSA test would be
remedied. (2) For the tissue subgroup, because of the level of sensitivity (0.84/0.87) and specificity (0.91/0.89) in two analyses, we suggest that the APC methylation test might be a better
test to distinguish PCa among the tissue. Although the biopsy was the gold standard in diagnosing PCa, there were still some errors when we did not obtain the cancer tissue well. With the
high sensitivity and specificity, the APC methylation test could be used to complement the biopsy, which would decrease the rate of FNs. (3) It has been said that methylation genes might
help identify new targets in the individual treatment of some diseases,55 and our study may provide stronger evidence of the potential function of these genes in finding a cure for PCa. In
addition, although in our analysis we did not find any evidence of publication bias, we should not ignore the potential bias that could affect the results. Nevertheless, regardless of the
possible bias, we suggest that studies of the function of methylation in identifying and curing the cancers cannot be neglected. LIMITATIONS The present study has several limitations. First,
the validation assay of the gene promoter methylation used in each study was different (MSP/QMSP/other). In addition, primers selected from different regions of the same CpG Island may have
different sensitivities and specificities (Table 1). Second, the thresholds identified by the ROC and INM were determined from individual trials, which may lead to different definitions of
methylation. Third, the sample collection time varied widely among the studies. Finally, as mentioned above, we analyzed associations between gene methylation and pathological stage, Gleason
score, PSA levels, and other factors, thereby decreasing our statistical power due to multiple testing. CONCLUSION PCa is a worldwide disease that affects a large number of men and leads to
a serious conclusion. To diagnose and interpose this disease early may indicate a good prognosis. Although the PSA test has been applied in disease diagnosis, its poor specificity limits
its function. On the basis of the studies available, this meta-analysis has demonstrated that APC methylation might be an ideal biomarker for screening and identifying PCa when combined with
the PSA test to decrease the rate of unnecessary biopsy. However, given the heterogeneity between the studies and insufficient evidence, the real function of APC methylation in disease
diagnosis requires further research. REFERENCES * Parkin DM, Bray F, Ferlay J _et al_: Global cancer statistics, 2002. _CA Cancer J Clin_ 2005; 55: 174–108. Article Google Scholar * Jemal
A, Siegel R, Ward E _et al_: Cancer statistics, 2008. _CA Cancer J Clin_ 2008; 58: 71–96. Article Google Scholar * Jemal A, Siegel R, Ward E _et al_: Cancer statistics, 2009. _CA Cancer J
Clin_ 2009; 59: 225–249. Article Google Scholar * Jemal A, Siegel R, Ward E _et al_: Cancer statistics, 2006. _CA Cancer J Clin_ 2006; 56: 106–130. Article Google Scholar * Zaridze DG,
Boyle P, Smans M : International trends in prostatic cancer. _Int J Cancer_ 1984; 33: 223–230. Article CAS Google Scholar * Cooner WH, Mosley BR, Rutherford CL _et al_: Prostate cancer
detection in a clinical urological practice by ultrasonography, digital rectal examination and prostate specific antigen. 1990. _J Urol_ 2002; 167: 966–973. Article Google Scholar * Stamey
TA, Caldwell M, McNeal JE _et al_: The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? _J Urol_ 2004; 172: 1297–1301.
Article Google Scholar * Eads CA, Danenberg KD, Kawakami K _et al_: MethyLight: a high-throughput assay to measure DNA methylation. _Nucleic Acids Res_ 2000; 28: E32. Article CAS Google
Scholar * Cho NY, Kim JH, Moon KC _et al_: Genomic hypomethylation and CpG island hypermethylation in prostatic intraepithelial neoplasm. _Virchows Arch_ 2009; 454: 17–23. Article CAS
Google Scholar * Costa VL, Henrique R, Jerónimo C : Epigenetic markers for molecular detection of prostate cancer. _Dis Markers_ 2007; 23: 31–41. Article CAS Google Scholar * Hoque MO,
Topaloglu O, Begum S _et al_: Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. _J Clin
Oncol_ 2005; 23: 6569–6575. Article CAS Google Scholar * Rogers CG, Gonzalgo ML, Yan G _et al_: High concordance of gene methylation in post-digital rectal examination and post-biopsy
urine samples for prostate cancer detection. _J Urol_ 2006; 176: 2280–2284. Article CAS Google Scholar * Rouprêt M, Hupertan V, Catto JW _et al_: Promoter hypermethylation in circulating
blood cells identifies prostate cancer progression. _Int J Cancer_ 2008; 122: 952–956. Article Google Scholar * Rouprêt M, Hupertan V, Yates DR _et al_: Molecular detection of localized
prostate cancer using quantitative methylation-specific PCR on urinary cells obtained following prostate massage. _Clin Cancer Res_ 2007; 13: 1720–1725. Article Google Scholar * Vener T,
Derecho C, Baden J _et al_: Development of a multiplexed urine assay for prostate cancer diagnosis. _Clin Chem_ 2008; 54: 874–882. Article CAS Google Scholar * Yoon HY, Kim SK, Kim YW _et
al_: Combined hypermethylation of APC and GSTP1 as a Molecular marker for prostate cancer: quantitative pyrosequencing analysis. _J Biomol Screen_ 2012; 17: 987–992. Article Google Scholar
* Kang GH, Lee S, Lee HJ _et al_: Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. _J Pathol_ 2004; 202: 233–240. Article
CAS Google Scholar * Bastian PJ, Ellinger J, Heukamp LC _et al_: Prognostic value of CpG island hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in patients undergoing
radical prostatectomy. _Eur Urol_ 2007; 51: 665–674. Article CAS Google Scholar * Ellinger J, Bastian PJ, Jurgan T _et al_: CpG island hypermethylation at multiple gene sites in diagnosis
and prognosis of prostate cancer. _Urology_ 2008; 71: 161–167. Article Google Scholar * Maruyama R, Toyooka S, Toyooka KO _et al_: Aberrant promoter methylation profile of prostate
cancers and its relationship to clinicopathological features. _Clin Cancer Res_ 2002; 8: 514–519. CAS PubMed Google Scholar * Jerónimo C, Henrique R, Hoque MO _et al_: A Quantitative
promoter methylation profile of prostate cancer. _Clin Cancer Res_ 2004; 10: 8472–8478. Article Google Scholar * Tokumaru Y, Harden SV, Sun DI _et al_: Optimal use of a panel of
methylation markers with GSTP1 hypermethylation in the diagnosis of prostate adenocarcinoma. _Clin Cancer Res_ 2004; 10: 5518–5522. Article CAS Google Scholar * Bastian PJ, Ellinger J,
Wellmann A _et al_: Diagnostic and prognostic information in prostate cancer with the help of a small set of hypermethylated gene loci. _Clin Cancer Res_ 2005; 11: 4097–4106. Article CAS
Google Scholar * Enokida H, Shiina H, Urakami S _et al_: Multigene methylation analysis for detection and staging of prostate cancer. _Clin Cancer Res_ 2005; 11: 6582–6588. Article CAS
Google Scholar * Henrique R, Jerónimo C, Teixeira MR _et al_: Epigenetic heterogeneity of high-grade prostatic intraepithelial neoplasia: clues for clonal progression in prostate
carcinogenesis. _Mol Cancer Res_ 2006; 4: 1–8. Article CAS Google Scholar * Yegnasubramanian S, Kowalski J, Gonzalgo ML _et al_: Hypermethylation of CpG islands in primary and metastatic
human prostate cancer. _Cancer Res_ 2004; 64: 1975–1986. Article CAS Google Scholar * Cho NY, Kim BH, Choi M _et al_: Hypermethylation of CpG island loci and hypomethylation of LINE-1 and
Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. _J Pathol_ 2007; 211: 269–277. Article CAS Google Scholar * Vasiljević N, Wu K, Brentnall
AR _et al_: Absolute quantitation of DNA methylation of 28 candidate genes in prostate cancer using pyrosequencing. _Dis Markers_ 2011; 30: 151–161. Article Google Scholar * Trock BJ,
Brotzman MJ, Mangold LA _et al_: Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high-risk cohort of men with negative initial prostate biopsies. _BJU Int_ 2012;
110: 56–62. Article CAS Google Scholar * Lau J, Ioannidis JP, Schmid CH : Quantitative synthesis in systematic reviews. _Ann Intern Med_ 1997; 127: 820–826. Article CAS Google Scholar
* Midgette AS, Stukel TA, Littenberg B : A meta-analytic method for summarizing diagnostic test performances: receiver-operating-characteristic-summary point estimates. _Med Decis Making_
1993; 13: 253–257. Article CAS Google Scholar * Wu T, Giovannucci E, Welge J _et al_: Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: a
meta-analysis. _Br J Cancer_ 2011; 105: 65–73. Article CAS Google Scholar * Schwarzenbach H, Chun FK, Isbarn H _et al_: Genomic profiling of cell-free DNA in blood and bone marrow of
prostate cancer patients. _J Cancer Res Clin Oncol_ 2011; 137: 811–819. Article CAS Google Scholar * Okegawa T, Nutahara K, Higashihara E : Association of circulating tumor cells with
tumor-related methylated DNA in patients with hormone-refractory prostate cancer. _Int J Urol_ 2010; 17: 466–475. Article CAS Google Scholar * Phé V, Cussenot O, Rouprêt M : Interest of
methylated genes as biomarkers in urothelial cell carcinomas of the urinary tract. _BJU Int_ 2009; 104: 896–901. Article Google Scholar * Phé V, Cussenot O, Rouprêt M : Methylated genes as
potential biomarkers in prostate cancer. _BJU Int_ 2010; 105: 1364–1370. Article Google Scholar * Baden J, Green G, Painter J _et al_: Multicenter evaluation of an investigational
prostate cancer methylation assay. _J Urol_ 2009; 182: 1186–1193. Article CAS Google Scholar * Baden J, Adams S, Astacio T _et al_: Predicting prostate biopsy result in men with prostate
specific antigen 2.0 to 10.0 ng/ml using an investigational prostate cancer methylation assay. _J Urol_ 2011; 186: 2101–2106. Article Google Scholar * Florl AR, Steinhoff C, Müller M _et
al_: Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. _Br J Cancer_ 2004; 91: 985–994. Article CAS Google Scholar * Van Neste L, Bigley
J, Toll A _et al_: A tissue biopsy-based epigenetic multiplex PCR assay for prostate cancer detection. _BMC Urol_ 2012; 12: 16. Article CAS Google Scholar * Stewart GD, Van Neste L,
Delvenne P _et al_: Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC Study. _J Urol_ 2012; S0022-5347:
04906–3. Google Scholar * Richiardi L, Fiano V, Vizzini L _et al_: Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. _J Clin Oncol_ 2009; 27:
3161–3168. Article CAS Google Scholar * Padar A, Sathyanarayana UG, Suzuki M _et al_: Inactivation of cyclin D2 gene in prostate cancers by aberrant promoter methylation. _Clin Cancer
Res_ 2003; 9: 4730–4734. CAS PubMed Google Scholar * Costa VL, Henrique R, Ribeiro FR _et al_: Epigenetic regulation of Wnt signaling pathway in urological cancer. _Epigenetics_ 2010; 5:
343–351. Article CAS Google Scholar * Liu L, Kron KJ, Pethe VV _et al_: Association of tissue promoter methylation levels of APC, TGFβ2, HOXD3 and RASSF1A with prostate cancer
progression. _Int J Cancer_ 2011; 129: 2454–2462. Article CAS Google Scholar * Steiner I, Jung K, Schatz P _et al_: Gene promoter methylation and its potential relevance in early prostate
cancer diagnosis. _Pathobiology_ 2010; 77: 260–266. Article CAS Google Scholar * Troyer DA, Lucia MS, de Bruïne AP _et al_: Prostate cancer detected by methylated gene markers in
histopathologically cancer-negative tissues from men with subsequent positive biopsies. _Cancer Epidemiol Biomarkers Prev_ 2009; 18: 2717–2722. Article CAS Google Scholar * Gillio-Tos A,
Fiano V, Zugna D _et al_: DNA methyltransferase 3b (DNMT3b), tumor tissue DNA methylation, Gleason score, and prostate cancer mortality: investigating causal relationships. _Cancer Causes
Control_ 2012; 23: 1549–1555. Article Google Scholar * Rosenbaum E, Hoque MO, Cohen Y _et al_: Promoter hypermethylation as an independent prognostic factor for relapse in patients with
prostate cancer following radical prostatectomy. _Clin Cancer Res_ 2005; 11: 8321–8325. Article CAS Google Scholar * Delgado-Cruzata L, Hruby GW, Gonzalez K _et al_: DNA methylation
changes correlate with Gleason score and tumor stage in prostate cancer. _DNA Cell Biol_ 2012; 31: 187–192. Article CAS Google Scholar * Henrique R, Ribeiro FR, Fonseca D _et al_: High
promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. _Clin Cancer Res_ 2007; 13: 6122–6129. Article CAS Google Scholar * Clark JP,
Munson KW, Gu JW _et al_: Performance of a single assay for both type III and type VI TMPRSS2:ERG fusions in noninvasive prediction of prostate biopsy outcome. _Clin Chem_ 2008; 54:
2007–2017. Article CAS Google Scholar * Neuhausen A, Florl AR, Grimm MO _et al_: DNA methylation alterations in urothelial carcinoma. _Cancer Biol Ther_ 2006; 5: 993–1001. Article CAS
Google Scholar * Catalona WJ : Management of cancer of the prostate. _N Engl J Med_ 1994; 331: 996–1004. Article CAS Google Scholar * Mahoney SE, Yao Z, Keyes CC _et al_: Genome-wide DNA
methylation studies suggest distinct DNA methylation patterns in pediatric embryonal and alveolar rhabdomyosarcomas. _Epigenetics_ 2012; 7: 400–408. Article CAS Google Scholar Download
references ACKNOWLEDGEMENTS This study was supported by grants from the Key Opening laboratory subject of Guangxi medical science experiment center (KFJJ2011-22). DISCLAIMER The funders had
no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. AUTHOR INFORMATION Author notes * Yang Chen, Jie Li and Xiaoxiang Yu: These
authors contributed equally to this work AUTHORS AND AFFILIATIONS * Center for Genomic and Personalized Medicine, Guangxi Medical University, Nanning, China Yang Chen, Jie Li, Shuai Li,
Zengnan Mo & Yanling Hu * Institute of Urology and Nephrology, the People's Liberation Army 303 Hospital of Guangxi, Nanning, China Xiaoxiang Yu * Medical Research Center, Guangxi
Medical University, Nanning, China Xuerong Zhang & Yanling Hu * Institute of Urology and Nephrology, First Affiliated Hospital of Guangxi Medical University, Nanning, China Zengnan Mo
Authors * Yang Chen View author publications You can also search for this author inPubMed Google Scholar * Jie Li View author publications You can also search for this author inPubMed Google
Scholar * Xiaoxiang Yu View author publications You can also search for this author inPubMed Google Scholar * Shuai Li View author publications You can also search for this author inPubMed
Google Scholar * Xuerong Zhang View author publications You can also search for this author inPubMed Google Scholar * Zengnan Mo View author publications You can also search for this author
inPubMed Google Scholar * Yanling Hu View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Zengnan Mo or Yanling Hu.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no confict of interest. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Chen, Y., Li, J., Yu,
X. _et al._ _APC_ gene hypermethylation and prostate cancer: a systematic review and meta-analysis. _Eur J Hum Genet_ 21, 929–935 (2013). https://doi.org/10.1038/ejhg.2012.281 Download
citation * Received: 23 August 2012 * Revised: 09 November 2012 * Accepted: 21 November 2012 * Published: 09 January 2013 * Issue Date: September 2013 * DOI:
https://doi.org/10.1038/ejhg.2012.281 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * hypermethylation * methylation * prostate cancer *
adenomatous polyposis coli
