Making a living while starving in the dark: metagenomic insights into the energy dynamics of a carbonate cave
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Carbonate caves represent subterranean ecosystems that are largely devoid of phototrophic primary production. In semiarid and arid regions, allochthonous organic carbon inputs
entering caves with vadose-zone drip water are minimal, creating highly oligotrophic conditions; however, past research indicates that carbonate speleothem surfaces in these caves support
diverse, predominantly heterotrophic prokaryotic communities. The current study applied a metagenomic approach to elucidate the community structure and potential energy dynamics of microbial
communities, colonizing speleothem surfaces in Kartchner Caverns, a carbonate cave in semiarid, southeastern Arizona, USA. Manual inspection of a speleothem metagenome revealed a community
genetically adapted to low-nutrient conditions with indications that a nitrogen-based primary production strategy is probable, including contributions from both _Archaea_ and _Bacteria_.
Genes for all six known CO2-fixation pathways were detected in the metagenome and RuBisCo genes representative of the Calvin–Benson–Bassham cycle were over-represented in Kartchner
speleothem metagenomes relative to bulk soil, rhizosphere soil and deep-ocean communities. Intriguingly, quantitative PCR found _Archaea_ to be significantly more abundant in the cave
communities than in soils above the cave. MEtaGenome ANalyzer (MEGAN) analysis of speleothem metagenome sequence reads found _Thaumarchaeota_ to be the third most abundant phylum in the
community, and identified taxonomic associations to this phylum for indicator genes representative of multiple CO2-fixation pathways. The results revealed that this oligotrophic subterranean
environment supports a unique chemoautotrophic microbial community with potentially novel nutrient cycling strategies. These strategies may provide key insights into other ecosystems
dominated by oligotrophy, including aphotic subsurface soils or aquifers and photic systems such as arid deserts. SIMILAR CONTENT BEING VIEWED BY OTHERS ACTIVE LITHOAUTOTROPHIC AND
METHANE-OXIDIZING MICROBIAL COMMUNITY IN AN ANOXIC, SUB-ZERO, AND HYPERSALINE HIGH ARCTIC SPRING Article 08 April 2022 DEPTHWISE MICROBIOME AND ISOTOPIC PROFILING OF A MODERATELY SALINE
MICROBIAL MAT IN A SOLAR SALTERN Article Open access 26 November 2020 NICHE DIFFERENTIATION OF SULFUR-OXIDIZING BACTERIA (SUP05) IN SUBMARINE HYDROTHERMAL PLUMES Article Open access 26
January 2022 INTRODUCTION Photosynthesis is the primary driver of energy dynamics in terrestrial and marine ecosystems, where light energy is harnessed for the conversion of atmospheric CO2
to reduced carbon. Avenues for understanding alternate non-photosynthetic primary production strategies are limited to subterranean or deep-sea ecosystems that function in the absence of
sunlight. A significant amount of research has been devoted to characterizing primary production in ecosystems such as hydrothermal vents (reviewed by Nakagawa and Takai, 2008) and sulfidic
caves (Sarbu et al., 1996; Chen et al., 2009; Engel et al., 2010; Jones et al., 2012), where supplies of reduced sulfur, hydrogen or methane support rich chemolithoautotophic activity;
however, the energy dynamics of carbonate caves are less well defined. Carbonate cave communities are presumed to be sustained by allocthonous carbon sourced from photic surface ecosystems
and entering the cave with vadose-zone drip water, surface water flow or the behavior of macrofauna (Laiz et al., 1999; Simon et al., 2003; Barton et al., 2004). In contrast, limited-access
carbonate caves in semiarid and arid regions are highly oligotrophic owing to low carbon levels in surface soils, low mean annual rainfall and small openings that prevent large scale
macrofauna exchange with surface ecosystems. As such, these caves provide a window for analyzing the metabolic flexibililty of microbial communities in an aphotic oligotrophic habitat with
potential similarity to diverse globally dominant terrestrial and marine environments, including subsoil to bedrock layers, oligotrophic aquifers and the deep ocean. Energy dynamics
elucidated from these aphotic ecosystems may also be applicable to comparably oligotrophic photic systems such as arid deserts, glacial and polar ice, and even extraterrestrial planetary
subsurface environments. In addition, analysis of the energy dynamics of carbonate caves provides information concerning the potential influences of microbial activity on carbon
sequestration in speleothems, the secondary carbonate deposits commonly found in caves. The latter application is particularly relevant to the widespread study of the isotopic composition of
speleothems for reconstruction of recent (Quaternary) climate change (Wang et al., 2005). Microbial contributions to speleothem isotopic signatures are not well understood, however,
research indicates that microbial activity enhances calcium carbonate precipitation (Contos et al., 2001; reviewed by Barton and Northup, 2007; Banks et al., 2010), and that carbon isotope
fractionation rates vary with different microbial CO2-fixation pathways (reviewed by Berg et al., 2010). Research characterizing the functional profiles of speleothem microbial communities
will enhance our understanding of the metabolic potential and energy dynamics of oligotrophic karst environments and provide critical information for applications such as the analysis of
speleothem isotopic signatures. Kartchner Caverns is a limited-access cave that developed within a Lower Carboniferous age Escabrosa limestone formation in the semiarid Whetstone Mountains
of southeastern Arizona, USA (Jagnow, 1999). The only natural entrance is a small blowhole, largely limiting macrofauna exchange to bats, insects and small rodents, amphibians and reptiles.
Human access has been tightly controlled since the beginning of cave development (Tufts and Tenen, 1999), and environmental conditions within the cave have been continuously monitored since
1989 (Toomey and Nolan, 2005). Current information on carbonate cave metabolic potential has been largely inferred from 16S rRNA gene molecular surveys rather than functional analyses (for
example, Barton et al., 2004, 2007; Ortiz et al., 2013). A broad pyrotag survey in Kartchner Caverns revealed an unexpectedly high bacterial diversity on speleothem surfaces with an average
of 1994 operational taxonomic units (OTUs) per speleothem that were classified in 21 phyla and 12 candidate phyla, and appeared to be dominated by organisms associated with heterotrophic
growth (Ortiz et al., 2013). Studies in other carbonate caves have proposed that cave microbes are translocated soil heterotrophs (Laiz et al., 1999; Simon et al., 2003) supported primarily
by allochthonous carbon compounds entering the cave with air currents or water percolating from the surface (Barton et al., 2004). In contrast, our pyrotag survey revealed an overlap of just
16% between cave OTUs and those of a soil community from above the cave. Phylogenetic associations to cave OTUs suggested the presence of chemolithoautotrophy within the cave. The objective
of this study was to examine the primary production metabolic capabilities of Kartchner Caverns speleothem communities using (1) an analysis of the functional and taxonomic composition of a
cave speleothem metagenome and (2) comparisons of the genomic profiles of multiple cave metagenomes to other terrestrial and marine environments to identify potentially important strategies
developed by carbonate cave microbes to survive the unique challenges of this oligotrophic, subterranean environment. These analyses were driven by the hypothesis that although speleothem
communities are dominated by heterotrophic bacteria, the source of energy for heterotrophs is at least partially derived from key chemolithoautotrophic activities. MATERIALS AND METHODS
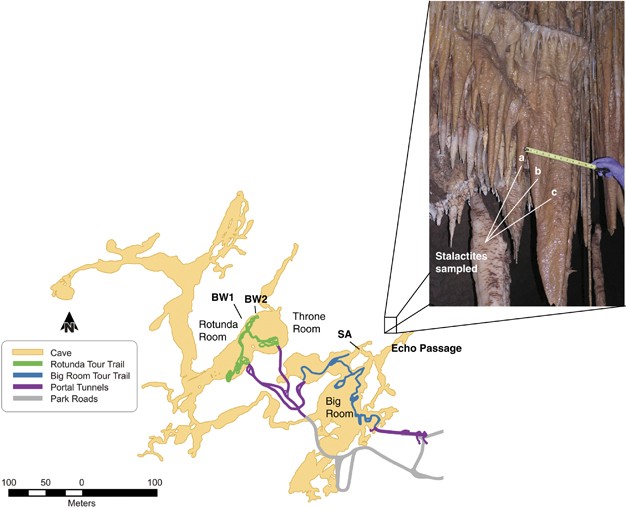
SAMPLING, DNA EXTRACTION AND SEQUENCING This study represents a metagenomic analysis of surface microbial communities sampled from a complex cave formation located in a remote room of
Kartchner Caverns accessed through the Echo Passage. The formation was sampled in October 2009 and consists of multiple stalactites descending from a common drapery (Figure 1). Thirty-two
swabs moistened with sterile deionized H2O were used to sample the surface area (4 cm2 per swab) of three stalactites from the Echo Passage speleothem using previously described protocols
(Ortiz et al., 2013). Previous research revealed variability in community composition along the surface of a single speleothem, but indicated that this variability is less than that observed
between distinct speleothems. (Legatzki et al., 2011). The composite sample created using 32 swabs to sample the majority of the surface area of three closely associated speleothems
descending from a single drapery was designed to incorporate potential variability in community composition along the speleothem surface. The swabs were sonicated (20 s), vortexed (1 min)
and sonicated again in sterile deionized H2O, and then removed. The remaining supernatant was centrifuged at 14 000_ g_ for 10 min. The resulting pellet was resuspended in 978 ml sodium
phosphate buffer and total genomic DNA was extracted using the FastDNA spin kit for soils (MP Biomedicals, Solon, OH, USA) following the manufacturer’s protocol optimized to enhance DNA
recovery from low template samples (Solis-Dominguez et al., 2011). Finally, 509 ng of DNA extract at a concentration of 9.8 ng μl−1 was provided to the Arizona Genomic Institute for
sequencing in one direction using the Rapid Library Preparation method for GS-FLX-Titanium pyrosequencing (454 Life Sciences, Roche Diagnostic Corporation, Branford, CT, USA). DRIP-WATER
COLLECTION Drip-water samples were collected monthly from January to December 2011 below a stalactite in the Big Wall room (Figure 1, BW2). Water was collected for a period of 5–23 days each
month until an average volume of 220 ml had been recovered. Dissolved organic carbon was measured following acidification to remove inorganic carbon, using a Shimadzu VCSH total organic
carbon analyzer (Columbia, MD, USA) with a solid state module (SSM-5000A). NO2-N and NO3-N was analyzed using a Dionex ICS-1000 (ThermoScientific, Sunnyvale, CA, USA). NO2-N was below the
detection limit in all samples. DATA PROCESSING, ASSEMBLY AND ANNOTATION Total sequence data obtained from the Echo Passage sample were 291 Mbp (930 939 reads of an average read length of
313 bp). Low quality reads were filtered from the data set based on the following criteria: a quality score two s.d. smaller than the average, a length two s.d. smaller or larger than the
average and ambiguous bases (Ns) (Huse et al., 2007; Hurwitz et al., 2012). The resulting 769 420 reads were then dereplicated (http://microbiomes.msu.edu/replicates; Gomez-Alvarez et al.,
2009), leaving 506 618 reads that were assembled using the Newbler software (454 Life Sciences, Roche Diagnostics Corporation) using a minimum length of 80 bp and a minimum sequence identity
of 96%. The assembled reads were submitted to the Integrated Microbial Genomes with Microbiomes Samples Expert Review (IMG/M ER) pipeline (Markowitz et al., 2009) for gene prediction and
annotation. In addition, the unassembled reads were analyzed by BLASTX (http://blast.ncbi.nlm.gov) against the NCBI non-redundant nucleotide (NR) database, then evaluated for taxonomic
assignments using the MEtaGenome ANalyzer (MEGAN v4.69.4) software with its default settings. MEGAN uses a lowest common denominator algorithm to assign reads to a taxa (Huson et al., 2007).
Details on in-house python scripts for analysis of large data sets and BLAST adaptive strategies are documented at: http://ag.arizona.edu/swes/maier_lab/kartchner/documentation/. The data
for the Echo Passage metagenome is available through the IMG/M database, IMG-ID 2189573024. Sequence data from two stalactites and one calcite-coated wet rock surface located below the Big
Wall in the Rotuda Room area of the cave (Figure 1, BW2) were generated by a concurrent study and were included for all comparative analyses. Sampling procedures, data processing, assembly
and annotation for the BW2 metagenomes followed the same protocols described above (Julia Neilson, personal communication). The three BW2 surfaces were comprehensively sampled with a total
of 60, 96 and 84 swabs, respectively, and produced DNA yields of 2900, 446 and 229 ng of DNA per surface. The average sequence read length of the three BW2 samples was 382–392 bp. The BW2
data is archived in the IMG/M database and will be released upon publication of the related manuscript. COMPARATIVE ANALYSIS Relative gene abundance was profiled by comparing the Echo
Passage cave metagenome to 15 other metagenomes (Supplementary Table 1) that included the three Kartchner Caverns BW2 cave metagenomes and 12 publicly available metagenomes obtained from the
IMG/M database (http://img.jgi.doe.gov/cgi-bin/m/main.cgi) that had been pre-processed (quality filter, assembly and so on) before submission to the IMG/M database. The twelve IMG/M
metagenomes included four from each of the following environments; deep ocean, bulk soil and rhizosphere soil. Gene prediction and annotation for all samples was done on assembled sequence
reads using the IMG/M ER pipeline. Hierarchical cluster analysis was performed using the IMG/M ER/Compare Genomes/Genome Clustering tool by function using the Clusters of Orthologous Groups
of proteins (COGs) classification system. The relative gene abundance for each metagenome sample was calculated for a given COG category/function by dividing the number of genes in that
category by the total number of genes with COG function assignment in order to reduce annotation bias (Delmont et al., 2011), and averages were generated for each environment. The COG
classification system was selected because it assigned functions to the largest number of gene sequences for each metagenome. Statistical significance was determined by one-way analysis of
variance (ANOVA) and the Tukey–Kramer honestly ignificant difference (HSD) test (_P_<0.05) as implemented in JMP9 (SAS Institute Inc., Cary, NC, USA). ANALYSIS OF SPECIFIC GENES The
metabolic analysis of the Echo Passage metagenome was performed using both the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the COGs classification systems. KEGG maps of carbon
fixation and nitrogen metabolism for the Echo Passage metagenome were obtained from the IMG/M ER pipeline. The Echo Passage protein sequences for specific enzymes involved in these two
processes were downloaded from the site and analyzed by BLASTP against the NCBI-NR database. BLAST results were then uploaded to MEGAN for taxonomic analysis and visualization using default
settings. QUANTITATIVE PCR ANALYSIS The relative abundance of bacteria, archaea and fungi present in the cave was compared with soil from above the cave by quantitative PCR (qPCR) using a
CFX96 Real-Time PCR System (Bio-Rad Laboratories, Hercules, CA, USA). The analysis was conducted using DNA extracts from four cave surfaces sampled near BW2 in Kartchner Caverns (Figure 1).
The surfaces included a dry rock wall (DRW), a wet rock coated with calcite veneer (WRW) and two speleothems (A and B). Soil samples were collected at a depth of 15–25 cm from three sites
above the cave including near the cave sink hole next to the natural cave entrance (SH), above the Rotunda Room portion of the cave (OR) and next to the rain gauge (RG) in the saddle between
the two hills over the cave. The soils were collected and processed as described by Drees et al. (2006). Soil cover above the cave is minimal being classified as primarily exposed bedrock
surfaces with intermittent pockets of soil development typically <1 m in depth. The sample depth was selected to avoid the influence of eolian deposition of surface materials while
preserving the rhizosphere influence of the photic surface ecosystem. Total genomic DNA from cave and soil samples was extracted using the FastDNA Spin Kit for soil and amplified in a 10-μl
qPCR reaction with 1 × SsoFast EvaGreen Supermix (Bio-Rad Laboratories), 400 μg ml−1 unacetylated bovine serum albumin solution, 400 nM of each primer and 400 pg DNA extract. Amplifications
used 16S rDNA primers 338F/518R and 931F/1100R for bacteria and archaea, respectively, (Einen et al., 2008) and 18S rDNA primers nu-SSU-1196F/nu-SSU-1536R for fungi (Borneman and Hartin,
2000; Castro et al., 2010). Standards for each domain were made from linearized plasmids (pGEM- T Easy; Promega, Madison, WI, USA) containing the SSU rRNA gene fragments from _Escherichia
coli_ JM109, _Halogeometricum borinquense_ ATCC 700274 and _Alternaria alternata_ for bacteria, archaea and fungi, respectively. The qPCR amplification conditions were: 98 °C for 3 min,
followed by 45 cycles of 98 °C for 5 s and 60 °C for 5 s (6 s for fungal qPCR). Sample traces were considered quantifiable if they fell within the range of reproducible standard traces for
the respective standard curves. Otherwise, the SSU rRNA gene was labeled undetectable. The following controls were included in all reactions. The positive control used DNA extracted from the
surface of an above-ground rock using the same swab and extraction protocols used on cave surfaces. Good amplification of bacteria, archaea and fungi was consistently observed from the
positive control. No template-negative controls were run for each domain, and values obtained were consistently below the range of the standard curves confirming the absence of significant
influence from reagent contaminants. Technical triplicates were averaged for each sample. Potential inhibition was evaluated using plasmid puc18 added to all DNA extracts, and was amplified
with the primers M13F and M13R. No inhibition was detected. Statistical significance was determined with a two-tailed _t_-test using the JMP9 software (SAS Institute Inc.). RESULTS ANALYSIS
OF DRIP-WATER CHEMISTRY Drip-water flow rates fluctuated during the year with the highest rates tracking the winter rains and the summer monsoons (Figure 2). Nitrate levels consistently
exceeded total dissolved organic carbon concentrations, and peak concentrations for both carbon and nitrogen predictably followed the Sonoran Desert spring bloom, which typically occurs
during March and April. A nitrate peak was also observed in October that was not accompanied by a parallel increase in total dissolved organic carbon. ECHO PASSAGE METAGENOME OVERVIEW AND
TAXONOMIC COMPOSITION The analysis of assembled reads from the Echo Passage speleothem metagenome (IMG/M ER pipeline) resulted in 365 407 predicted genes of which ∼50% had predicted known
functions (Table 1). Based on the COGs classification system, the most abundant metabolic categories represented were amino acid transport and metabolism (10%), energy production and
conversion (8%), and replication, recombination and repair (7%) (Supplementary Figure 1). Analysis using the KEGG database also showed amino acid and energy metabolisms among the most
abundant categories (Supplementary Figure 2). Taxonomic analysis of the unassembled reads using a combination of MEGAN software and the NCBI-NR database produced classifications for 69% of
the reads (Figure 3). Of those classified, _Bacteria_ were dominant (85%) followed by _Archaea_ (10%), _Eukaryota_ (5%) and viruses (0.13%). Within the bacterial domain, 54% of the sequences
could be further assigned to a phylum and these were dominated by _Proteobacteria_ (52%), _Actinobacteria_ (13%) and _Planctomycetes_ (7.5%). This bacterial taxonomic distribution supports
recent pyrotag and clone analyses performed in two other regions of Kartchner Caverns (Figure 1, BW1 and SA) that also found _Protoebacteria_ and _Actinobacteria_ to be the dominant phyla
(Legatzki et al., 2011; Ortiz et al., 2013). However, the specific Echo Passage speleothem taxonomic profile was distinct from the nine other previously characterized speleothems, supporting
our previous observation that bacterial community composition varies among speleothems. MEGAN assigned 82% of the archaeal sequences to phyla (Figure 3), and these were dominated by
_Thaumarchaeota_ (76%) followed by _Euryarchaeota_ (17%) and _Crenarchaeota_ (7.6%). The majority of the _Thaumarchaeota_ sequences (53%) were associated with marine archaea. Further
analysis revealed that 13% of the _Thaumarchaeota_ reads were classified as the _Nitrosopumilus maritimus_, an ammonia-oxidizing marine archaeon (member of group I.1a) (Konneke et al.,
2005), and 4.7% were associated with _Nitrososphaera gargensis_ (member of group I.1b), an ammonia-oxidizer frequently found in terrestrial environments (Hatzenpichler et al., 2008). Within
the _Eukaryota_, 79% of the sequences could be classified of which 23% were fungi, accounting for just 0.9% of the total classified community. Other significant members of the classified
eukaryotic community include _Metazoa_ (37%), _Viridiplantae_ (16%) and _Alveolata_ (5%). Domain distributions were further evaluated by SSU rDNA qPCR, for comparison with metagenome results
and soils above the cave (Figure 4). As with the metagenome analysis, the qPCR results showed the cave communities to be dominated by _Bacteri_a followed by _Archaea_. Fungal genes were
below the detection level. Fungal genes amplified from the rock used as a positive control (see Materials and methods) were more abundant than those present in the soils, confirming that the
absence of fungal genes on cave surfaces was not an artifact of the sampling procedure or the primers used. Bacterial abundance in cave communities was comparable to the soil samples,
however, archaeal abundance was significantly higher in the cave than in the soil communities. In contrast, fungal abundance was significantly greater in the soil communities than on cave
surfaces. COMPARATIVE METAGENOMIC ANALYSIS A comparative metagenomic analysis of the assembled reads was performed using 4 Kartchner cave metagenomes and 12 publically available metagenomes
from deep-ocean, bulk soil and rhizosphere soil samples as explained in Materials and methods (Supplementary Table 1). These metagenome groups will be referred to as cave, ocean, soil and
rhizosphere, respectively, for the remainder of this paper. Hierarchical cluster analysis grouped the four cave samples in a separate clade more closely associated with soil and rhizosphere
samples than with ocean samples (Figure 5). The cluster analysis suggests that the cave communities represent distinct and potentially specialized terrestrial microbial communities. Over-
and under-representation of cave COG categories were evaluated by comparison with soil, rhizosphere and ocean environments. COG categories were classified as similar if no significant
difference was found between the cave and any of the ocean, soil or rhizosphere habitats (Figure 6a), or as variable if the cave was significantly different from at least one of the other
three habitats (_P_<0.05; Figure 6b). Results showed a significant difference between the cave communities and at least one of the other three habitats (ocean, soil or rhizosphere) for
50% of the COG categories (Figure 6b). Among these variable categories, significant over-representation in the cave communities was observed for replication, recombination and repair (L).
The over-represented genes in this category were primarily associated with uncharacterized proteins involved in DNA repair (COGs 1336, 1337, 1343, 1367 and 1518). Under-representation in
cave communities was found for carbohydrate transport and metabolism (G), though a significant difference was only found when comparing the cave with soil and rhizosphere metagenomes.
Specifically, differences were observed for genes associated with a monosaccharide ABC-transporter; COGs 1129, 1879 and 1869 were significantly lower in the cave and ocean compared with the
soil and rhizosphere metagenomes. These three genes together with COG1172, which was significantly under-represented in the cave relative to all three environments, represent the four
components of the ribose ABC-transporter, a two-component system involved in importing nutrients into the cell. Under-representation of this transporter likely reflects the low-nutrient
availability of this ecosystem (Lauro et al., 2009). Within carbohydrate metabolism, all genes for major pathways such as glycolysis, pentose phosphate pathway and the Entner–Doudoroff
pathway were detected in the Echo Passage metagenome. Specific patterns that differentiated the oligotrophic ecosystems (cave and oceans) from the typically richer ecosystems (soil and
rhizosphere) were also analyzed. The signal transduction (T) and defense mechanism (V) COGs were found to be significantly lower in the oligotrophic environments than in the richer ones.
Over-represented categories included coenzyme transport and metabolism (H), translation, ribosomal structure and biogenesis (J), and post-translational modification, protein turnover and
chaperones (O). Transcription (K) represented the only cave COG with abundance more similar to the soil and rhizosphere samples than to the ocean (Figure 6b). The covariance of the
oligotrophic ecosystems was of particular interest given that the cluster analysis indicated that the cave communities were generally more similar to both soil and rhizosphere than to the
ocean metagenomes. CAVE ENERGY AND NUTRIENT DYNAMICS INFERRED FROM THE ECHO PASSAGE METAGENOME CO2 FIXATION Potential primary production strategies in Kartchner Caverns were of primary
interest because of the low total dissolved organic carbon levels in cave drip water (Figure 2). KEGG analysis of the Echo Passage metagenome identified putative genes from all six known
CO2-fixation pathways in the metagenome (Supplementary Figure 3A and 3B), however, only the Calvin–Benson–Bassham (CBB) and the Arnon–Buchanan reverse tricarboxylic acid (rTCA) cycles were
fully represented. Putative RuBisCO genes (representing the CBB cycle, _n_=22) identified in the Echo Passage metagenome were further analyzed for taxonomic identification using MEGAN. Genes
belonging to both _Bacteria_ and _Archaea_ were identified and the relative abundance of those with domain assignments (50%) was 10:1 (_Bacteria_ to _Archaea_). Just 14% could be classified
below the domain level using MEGAN (Supplementary Figure 4A); therefore, BLASTP searches against the NCBI-NR database were conducted with the unidentified genes to obtain further taxonomic
associations. Top hits indicated that 45% of the RuBisCO genes in the Echo Passage metagenome were associated with _Proteobacteria_. Genus level associations were only obtained for the
betaproteobacterial genus _Nitrosospira_ (82% amino acid sequence identity) and the actinobacterium _Acidithiomicrobium_ (84% identity), both of which are known to fix CO2 using the CBB
cycle (Utaker et al., 2002; Norris et al., 2011). Seventy-nine putative genes corresponding to the key rTCA cycle indicator gene, ATP–citrate lyase, were also classified with MEGAN
(Supplementary Figure 4B) and 89% could be assigned to a domain with a 6:1 ratio of _Bacteria_ to _Archaea_. Of the bacterial genes, 44% could be assigned to _Proteobacteria_ (_n_=27), 11.5%
to _Nitrospirae_ (_n_=7), 11.5% to _Actinobacteria_ (_n_=7) and 8% to _Chloroflexi_ (_n_=5). Of these phyla, previous research has documented autotrophic growth by both _Proteobacteria_ and
_Nitrospirae_ using the rTCA cycle (Berg, 2011). The assignment of nine ATP–citrate lyase genes to the archaeal domain is of particular interest for future study because there is
conflicting evidence in the literature concerning the ability of archaea to assimilate CO2 using the rTCA cycle (Strauss et al., 1992; Ramos-Vera et al., 2009; Berg et al., 2010). Additional
insights into CO2-fixation mechanisms present in the Echo Passage community were obtained using the COG classification system. This analysis identified 130 putative genes for the aromatic
ring hydroxylase (COG2368), also known as 4-hydroxybutyryl-CoA dehydratase. For _Archaea_, 4-hydroxybutyryl-CoA dehydratase is considered as the key indicator enzyme in two archaeal
CO2-fixation mechanisms (Berg et al., 2010): the 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle (Berg et al., 2007) and the dicarboxylate-4-HB (DC/HB) cycle (Huber et al., 2008). This
gene is considered as a marker for CO2 fixation by _Thaumarchaeota_ (Zhang et al., 2010). In contrast, in bacteria, it is associated with fermentation rather than CO2 fixation (Gerhardt et
al., 2000). Taxonomic analysis using MEGAN could confirm 24% of these genes belonging to _Archaea_, 8.5% of which were _Thaumarchaeota_ (Supplementary Figure 4C). Not all the genes for the
HP/HB or the DC/HB cycles were identified in the Echo Passage metagenome, which is not surprising given the paucity of oligotrophic environmental isolates in current databases used for gene
annotation. The abundance of putative archaeal genes for 4-hydroxybutyryl-CoA dehydratase and ATP–citrate lyase supports the hypothesis that autotrophic archaea contribute to cave ecosystem
carbon assimilation. To further explore the relative importance of autotrophy in the cave, a COG-based comparative analysis of the CBB, rTCA and HP/HB-DC/HB indicator enzymes was performed
using the set of 16 metagenomes (soil, rhizosphere, cave and ocean) described previously (Figure 7). Statistical analysis showed significant over-representation of RuBisCO genes (COG1850,
CBB cycle) in the cave relative to the other three metagenomes. The ATP–citrate lyase (COG2301, rTCA) and the 4-hydroxybutyryl-CoA dehydratase (COG2368, HP/HB and DC/HB) were also more
abundant in both oligotrophic ecosystems (cave and ocean) than in the soil and in the rhizosphere, however, the differences were not significant due to the high variability between samples.
A KEGG-based analysis supported the COG results for RuBisCO and ATP–citrate lyase (not shown), but the 4-hydroxybutyryl-CoA dehydratase was not identified using the KEGG database. NITROGEN
METABOLISM The availability of reduced compounds as energy sources for primary production in carbonate caves is presumed to be limited to the host rock and drip water because of the absence
of specific energy sources (for example, reduced sulfur compounds present in sulfidic caves). Drip-water inorganic nitrogen appeared particularly relevant to the Kartchner Caverns ecosystem
because NO3-N concentrations in cave drip water consistently exceeded the dissolved organic carbon levels (Figure 2). An extensive analysis of 42 nitrogen cycling genes was performed
revealing a diversity of nitrification, nitrate reduction and ammonia assimilation genes in the Echo Passage speleothem metagenome (Supplementary Figure 5). Based on drip-water chemistry, we
were particularly interested in nitrification as a potential energy source for this oligotrophic cave ecosystem. Twenty-three ammonia monooxygenase (_amoA_) genes involved in the first step
of nitrification were detected in the Echo passage metagenome and were found using MEGAN classification to be equally distributed between the archaeal and bacterial domains (Supplementary
Figure 6). Genes for hydroxylamine oxidase, a key enzyme required for ammonia oxidation by bacteria (AOB), but not for ammonia-oxidizing archaea (AOA) (Hallam et al., 2006; Kim et al., 2011)
were not detected, suggesting that AOA are the predominant ammonia oxidizers in the cave ecosystem. Genes for nitrite oxidoreductase, responsible for the oxidation of nitrite to nitrate,
were not identified by gene annotation based on KEGG and COG databases. Nevertheless, BLASTP searches against the Echo Passage metagenome using the IMG/M ER BLAST tool revealed 62 Echo genes
with 30–97% identity (_E_-value<1e-5) to a putative nitrite oxidoreductase enzyme (YP_003798871) from the metagenome of _Candidatus Nitrospira defluvii_, a known nitrite oxidizer (Lucker
et al., 2010). Ammonia assimilation, denitrification and dissimilatory nitrate reduction pathways were all well represented in the Echo Passage metagenome (Supplementary Figure 5). In
addition, the marker gene (_nrfA_) (Kraft et al., 2011) for dissimilatory nitrate reduction to ammonium (DNRA) (Supplementary Figure 5) was detected. Comparative metagenomic analysis
revealed strong representation of COG0004 (ammonia permease), COG1251 (NAD(P) H-nitrite reductase large subunit) and COG2146 (nitrite reductase–ferredoxin) in the speleothem communities
(Figure 8). COG2146, involved in assimilatory reduction of nitrate and DNRA, was significantly over-represented in the cave metagenomes relative to the other environments. The
NAD(P)H-nitrite reductase large subunit is also associated with pathways for both assimilatory nitrate reduction and DNRA (KEGG). Finally, key genes for nitrogen fixation, including the
_nifD_ and _nifK_ genes, which encode the α- and β-subunits of the dinitrogenase enzyme, were not identified by the KEGG classification system. COGs analysis identified only the _nifH_ and
_nifF_ genes that encode for dinitrogenase reductase and flavodoxins, respectively. DISCUSSION The functional and taxonomic analyses presented in this study provide new insights into the
ecosystem dynamics and survival strategies of microbial communities colonizing speleothem and rock surfaces in the oligotrophic Kartchner Caverns habitat. In this study, we focused
specifically on surface communities because we believe them to be dynamic and most responsive to the on-going flux in drip-water nutrient inputs. Information from this study can inform
future work focused on microbial communities embedded in calcite formations that might provide a more long-term temporal archive of past environmental conditions and microbial influences on
calcite precipitation. The metagenomic analysis revealed that the cave microbial communities clustered apart from soil, rhizosphere and ocean environments, but were more similar to the soil
and rhizosphere metagenomes than to the ocean. Despite the closer association observed between the three terrestrial metagenome groups, a greater covariance of under- and over-represented
COG categories was observed for the two low-nutrient ecosystems (ocean and cave) than for the three terrestrial metagenomes. The two COG categories that were significantly under-represented
in both the cave and ocean metagenomes (T and V) were previously identified among genomic markers for confirming the trophic strategy of uncultured marine oligotrophs (Lauro et al., 2009).
In addition, our previous characterizations of Kartchner speleothem community diversity identified clones and cultured isolates with 99% and 98% identity, respectively, to classic marine
oligotrophs such as _Sphingopyxis alaskensis_ and _Polaromonas aquatica_ (Ikner et al., 2007; Ortiz et al., 2013), indicating that community members from the two habitats are
phylogenetically related, suggesting potential ecological similarities between these oligotrophic communities. Taken together, these results allow speculation that carbonate cave communities
originated from soil ecosystems, but that the specific low-nutrient environmental conditions of Kartchner Caverns have selected for oligotropic functional profiles that parallel trophic
strategies found in oligotrophic marine environments. ENERGY DYNAMICS OF THE ECHO PASSAGE MICROBIAL COMMUNITY Metagenomic analysis of the carbon and nitrogen metabolic pathways along with
the associated functional taxonomic composition provides strong evidence for a chemolithoautotrophic component to the Echo Passage speleothem microbial community. Full analysis required
creative use of multiple databases and analysis strategies to compensate for the limited representation of cave microbes and oligotrophic metagenomes in current databases. First, an
abundance of genes representing all six known CO2-fixation pathways was detected revealing the existence of a microbial community with diverse autotrophic potential. Putative RuBisCo genes
representing the CBB cycle were significantly over-represented relative to soil, rhizosphere and oceans, and the abundances of both ATP–citrate lyase (rTCA cycle) and 4-hydroxybutyryl-CoA
dehydratase (HP/HB-DC/HB cycles) genes were comparable to the nutrient-limited ocean environments and greater than both soil and rhizosphere. The abundance of HP/HB genes was of particular
interest because enzymes in this cycle use bicarbonate as the active inorganic carbon species, whereas bicarbonate is not a RuBisCo substrate. The HP/HB pathway is hypothesized to be
advantageous for chemolithoautotrophic marine archaea because bicarbonate availability under slightly alkaliphilic conditions (for example, ocean water) is significantly higher than
dissolved CO2 (Berg, 2011), conditions that may also apply to this carbonate cave ecosystem where drip water pH averages 8.0. The diversity of key genes representing the CBB, rTCA and HP/HB
pathways suggests the potential for community CO2 fixation under diverse environmental conditions. Each pathway has different energy demands as reviewed by Berg (2011) and demonstrated by
the example of a gammaproteobacterial marine endosymbiont that uses the relatively energy-expensive CBB cycle under high-energy conditions and the energetically more favorable rTCA cycle
under low-energy conditions (Markert et al., 2007). The majority of Echo Passage RuBisCo genes (CBB) were associated with _Proteobacteria_, the dominant phylum in the community. In contrast,
the ATP–citrate lyase genes for the rTCA cycle were assigned to a greater diversity of phyla including _Proteobacteria, Nitrospirae, Actinobacteria, Chloroflexi_ and the domain, _Archaea._
The chemolithoautotrophic _Nitrospirae_ exemplify chemoautotrophic K-strategists capable of growing on nitrite substrate concentrations 10-fold lower than that required by the
well-characterized _Nitrobacter_ species (Bartosch et al., 2002). Putative oxidoreductase genes with similarity to _C. Nitrospira defluvii_ were identified in the Echo metagenome, allowing
us to speculate that chemolithoautotrophic nitrite-oxidizing _Nitrospirae_ are key primary producers in this oligotrophic community. This hypothesis is supported by previous 454-pyrotag
surveys that found _Nitrospirae_ on all speleothem surfaces sampled within the cave (Ortiz et al., 2013). _Nitrospirae_ have also been identified globally in cave communities, including
Altamira Cave, Spain (Portillo et al., 2008; Porca et al., 2012), Niu Cave, China (Zhou et al., 2007), Pajsarjeva jama, Slovenia (Porca et al., 2012), Nullarbor caves, Australia (Holmes et
al., 2001; Tetu et al., 2013) and Spider and Lechuguilla caves, New Mexico (Northup et al., 2003). A second key insight offered by this work is the potential contribution of _Archaea_ to the
energy dynamics of this oligotrophic karst ecosystem. qPCR analysis revealed that archaeal abundance in Kartchner rock and speleothem communities is significantly higher than in soil
communities from above the cave. A recent global 16S rRNA pyrotag analysis found an average archaeal abundance in soils of 2% with the abundance inversely correlated with C:N ratio,
suggesting that _Archaea_ can tolerate or even exploit low-nutrient conditions (Bates et al., 2011). The taxonomic analysis of the Echo Passage metagenome found that unassembled archaeal
sequences comprised 10% of the metagenome. _Thaumarchaeota_ were dominant among archaea and represented the third most abundant phylum overall. Among the _Thaumarchaeota_, MEGAN found 18% of
the reads to be associated with ammonia-oxidizing archaea. Importantly, taxonomic associations to this phylum were also identified for both the ATP–citrate lyase (rTCA cycle) and
4-hydroxybutyryl-CoA dehydratase (HP/HB and DC/HB cycles) CO2-fixation genes. In addition, half of the _amoA_ genes identified in the Echo Passage metagenome were classified as _Archaea._
Previous studies have identified _Thaumarchaeota_ as chemoautotrophic ammonia oxidizers (Zhang et al., 2010; Pester et al., 2011) adapted to low substrate conditions (Martens-Habbena et al.,
2009). Further, AOA in soils have been shown to fix CO2 using the HP/HB cycle (Pratscher et al., 2011). Archaeal _amoA_ genes have also been amplified from a stalactite sample taken from a
mine adit in Colorado (Spear et al., 2007). Finally, a previous analysis of archaeal community structure on the SA speleothems in Kartchner Caverns (Figure 1) identified OTUs that were
phylogenetically associated with the AOA, _N. gargensis_ (Legatzki et al., 2011). Taken together, these results suggest that _Thaumarcheota_ represent a second key group of
chemolithoautotrophic primary producers in the Echo Passage speleothem community. A recent metagenomic analysis of microbial slime in the subterranean aquatic Weebubbie cave below
Australia’s Nullarbor Plain found a similar pattern of primary production driven by inorganic nitrogen metabolism (Tetu et al., 2013). Similar to Kartchner, Weebubbie microbial communities
included an abundance of ammonia-oxidizing _Thaumarchaeota_, although the Weebubbie community had a much higher relative abundance of _Thaumarchaeota,_ and the AOA:AOB ratio was more similar
to that found in marine environments. Thus, the Kartchner Caverns ecosystem again appears to represent an oligotrophic terrestrial counterpart to the energy dynamics observed in
oligotrophic marine habitats such as Weebubbie cave. The presence of archaea has been previously documented in numerous terrestrial carbonate caves (Northup et al., 2003; Chelius and Moore,
2004; Gonzalez et al., 2006; Macalady et al., 2007; Legatzki et al., 2011), but their physiological role has remained elusive. This metagenomic analysis strongly suggests that
_Thaumarchaeota_ represent a second group of nitrification-based primary producers in Kartchner Caverns. The data also indicate the potential involvement of _Archaea_ in alternate primary
production strategies due to the abundance of archaeal CO2-fixation genes not assigned specific taxonomic classifications. Molecular diversity and functional gene characterizations of Movile
Cave in Romania provide an intriguing contrast to these carbonate caves (Chen et al., 2009). Similar to Kartchner, Movile Cave is aphotic and sustained by chemolithoautotrophy, however, the
atmosphere in Movile is rich in hydrogen sulfide and methane, and supports high microbial productivity and rich fauna. Microbial mats sustained by sulfur- and ammonia oxidizers contain a
diversity of autotrophic bacterial phylotypes, but no archaea. The absence of archaea is in stark contrast to their abundance in Katchner, Weebubbie and the other carbonate caves listed
above. The contrast between these cave ecosystems suggests that low-nutrient carbonate caves provide a template for evaluating the role of archaea in oligotrophic terrestrial ecosystems with
potential application to subsurface soils, low-nutrient aquifers and even subsurface extraterrestrial ecosystems. A final cave ecosystem survival strategy highlighted by this study was the
over-representation of DNA repair enzyme genes. These enzymes belong to the RAMP (Repair Associated Mysterious Proteins) superfamily, a group of proteins for which no specific function is
known, but that clearly associate with DNA repair mechanisms. The overabundance of DNA repair genes is surprising given the absence of typical DNA-damaging agents such as ultraviolet light.
We hypothesize that the exceedingly high calcium concentrations in cave drip water (Legatzki et al., 2012) are the source of stress. Research in carbonate caves indicates that cave bacteria
precipitate calcium carbonate as a mechanism to overcome calcium toxicity (Banks et al., 2010). Similarly, previous work has linked high levels of calcium ions in eukaryotic cells with DNA
strand breakage (Cantoni et al., 1989). Specifically, it has been shown that when cells are under oxidative stress, calcium homeostasis is disrupted, leading to increases in intracellular
calcium concentrations that result in the activation of nucleases that damage DNA. Thus, we hypothesize that the abundance of DNA repair enzymes is an adaptation of cave microbes to the
stress caused by high calcium concentrations in the cave ecosystem. CONCLUSION This Echo Passage speleothem metagenome analysis combined with previously published pyrotag surveys provides
strong evidence for carbonate cave microbial communities specifically adapted to low nutrient and high calcium conditions, and most probably sustained at least in part by an inorganic
nitrogen-based primary production strategy with contributions from both bacteria and archaea. The diversity of CO2-fixation pathways represented in the Echo Passage metagenome suggests that
the Kartchner speleothem communities are primed to exploit the observed seasonal fluctuations in drip-water nutrient content, a hypothesis that can be tested in future temporal studies,
using qPCR to target the CO2-fixation genes identified in this work. In addition, unique nutrient conservation strategies, for example, DNRA, may be present such as suggested by the presence
of the _nrfA_ gene and the over-representation of COG2146 (nitrite reductase-ferrodoxin). Research has shown that DNRA capability is phylogenetically widespread (Kraft et al., 2011), is the
dominant or sole nitrate consumption pathway in a diversity of soils (reviewed in Rütting et al., 2011) and may represent a unique strategy of oligotrophic microbial communities to minimize
nitrogen loss. Despite numerous studies in terrestrial and aquatic systems, there is little consensus concerning the relative importance of this pathway to nitrogen dynamics or the specific
environmental factors that control DNRA activity in soils (Rütting et al., 2011). Finally, the metagenomic analysis of novel microbial communities is typically constrained by the limited
number of relevant organisms represented in the KEGG, COG and NCBI-NR databases. However, this analysis demonstrates that even with these limitations, key indicators of trophic dynamics in
Kartchner Caverns were identified that provide new insights into the primary production potential and survival strategies of this carbonate cave microbial community with potential relevance
to a diversity of terrestrial oligotrophic habitats. REFERENCES * Banks ED, Taylor NM, Gulley J, Lubbers BR, Giarrizo JG, Bullen HA _et al_ (2010). Bacterial calcium carbonate precipitation
in cave environments: a function of calcium homeostasis. _Geomicrobiol J_ 27: 444–454. Article CAS Google Scholar * Barton HA, Northup DE . (2007). Geomicrobiology in cave environments:
past, current and future perspectives. _J Cave Karst Stud_ 69: 163–178. Google Scholar * Barton HA, Taylor MR, Pace NR . (2004). Molecular phylogenetic analysis of a bacterial community in
an oligotrophic cave environment. _Geomicrobiol J_ 21: 11–20. Article CAS Google Scholar * Barton HA, Taylor NM, Kreate MP, Springer AC, Oehrle SA, Bertog JL . (2007). The impact of host
rock geochemistry on bacterial community structure in oligotrophic cave environments. _Int J Speleol_ 36: 93–104. Article Google Scholar * Bartosch S, Hartwig C, Spieck E, Bock E . (2002).
Immunological detection of _Nitrospira_-like bacteria in various soils. _Microb Ecol_ 43: 26–33. Article CAS PubMed Google Scholar * Bates ST, Berg-Lyons D, Caporaso JG, Walters WA,
Knight R, Fierer N . (2011). Examining the global distribution of dominant archaeal populations in soil. _ISME J_ 5: 908–917. Article CAS PubMed Google Scholar * Berg IA . (2011).
Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. _Appl Environ Microbiol_ 77: 1925–1936. Article CAS PubMed PubMed Central Google Scholar * Berg
IA, Kockelkorn D, Buckel W, Fuchs G . (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in archaea. _Science_ 318: 1782–1786. Article CAS
PubMed Google Scholar * Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hugler M _et al_ (2010). Autotrophic carbon fixation in archaea. _Nat Rev Microbiol_ 8: 447–460. Article
CAS PubMed Google Scholar * Borneman J, Hartin RJ . (2000). PCR primers that amplify fungal rRNA genes from environmental samples. _Appl Environ Microbiol_ 66: 4356–4360. Article CAS
PubMed PubMed Central Google Scholar * Cantoni O, Sestili P, Cattabeni F, Bellomo G, Pou S, Cohen M _et al_ (1989). Calcium chelator quin 2 prevents hydrogen-peroxide-induced DNA breakage
and cytotoxicity. _Eur J Biochem_ 182: 209–212. Article CAS PubMed Google Scholar * Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW . (2010). Soil microbial community responses to
multiple experimental climate change drivers. _Appl Environ Microbiol_ 76: 999–1007. Article CAS PubMed Google Scholar * Chelius M, Moore J . (2004). Molecular phylogenetic analysis of
archaea and bacteria in Wind Cave, South Dakota. _Geomicrobiol J_ 21: 123–134. Article CAS Google Scholar * Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H _et al_ (2009).
Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. _ISME J_ 3: 1093–1104. Article CAS PubMed Google Scholar * Contos AK,
James JM, Heywood K, Pitt K, Rogers P . (2001). Morphoanalysis of bacterially precipitated subaqueous calcium carbonate from Weebubbie Cave, Australia. _Geomicrobiol J_ 18: 331–343. Article
CAS Google Scholar * Delmont TO, Malandain C, Prestat E, Larose C, Monier JM, Simonet P _et al_ (2011). Metagenomic mining for microbiologists. _ISME J_ 5: 1837–1843. Article CAS
PubMed PubMed Central Google Scholar * Drees KP, Neilson JW, Betancourt JL, Quade J, Henderson DA, Pryor BM _et al_ (2006). Bacterial community structure in the hyperarid core of the
Atacama Desert, Chile. _Appl Environ Microbiol_ 72: 7902–7908. Article CAS PubMed PubMed Central Google Scholar * Einen J, Thorseth IH, Ovreas L . (2008). Enumeration of archaea and
bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy. _FEMS Microbiol Lett_ 282: 182–187. Article CAS PubMed Google Scholar * Engel AS, Meisinger DB,
Porter ML, Payn RA, Schmid M, Stern LA _et al_ (2010). Linking phylogenetic and functional diversity to nutrient spiraling in microbial mates from Lower Kane Cave (USA). _ISME J_ 4: 98–110.
Article PubMed Google Scholar * Gerhardt A, Cinkaya I, Linder D, Huisman G, Buckel W . (2000). Fermentation of 4-aminobutyrate by clostridium aminobutyricum: Cloning of two genes involved
in the formation and dehydration of 4-hydroxybutyryl-CoA. _Arch Microbiol_ 174: 189–199. Article CAS PubMed Google Scholar * Gomez-Alvarez V, Teal TK, Schmidt TM . (2009). Systematic
artifacts in metagenomes from complex microbial communities. _ISME J_ 3: 1314–1317. Article PubMed Google Scholar * Gonzalez JM, Portillo MC, Saiz-Jimenez C . (2006). Metabolically active
Crenarchaeota in Altamira Cave. _Naturwissenschaften_ 93: 42–45. Article CAS PubMed Google Scholar * Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J _et al_
(2006). Genomic analysis of the uncultivated marine crenarchaeote _Cenarchaeum symbiosum_. _Proc Natl Acad Sci USA_ 103: 18296–18301. Article CAS PubMed PubMed Central Google Scholar *
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H _et al_ (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. _Proc Natl Acad Sci USA_
105: 2134–2139. Article CAS PubMed PubMed Central Google Scholar * Holmes AJ, Tujula NA, Holley M, Contos A, James JM, Rogers P _et al_ (2001). Phylogenetic structure of unusual aquatic
microbial formations in Nullarbor Caves, Australia. _Environ Microbiol_ 3: 256–264. Article CAS PubMed Google Scholar * Huber H, Gallenberger M, Jahn U, Eylert E, Berg IA, Kockelkorn D
_et al_ (2008). A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic archaeum _Ignicoccus hospitalis_. _Proc Natl Acad Sci USA_ 105: 7851–7856.
Article CAS PubMed PubMed Central Google Scholar * Hurwitz BL, Deng L, Poulos BT, Sullivan MB . (2012). Evaluation of methods to concentrate and purify ocean virus communities through
comparative, replicated metagenomics. _Environ Microbiol_ 15: 1428–1440. Article PubMed Google Scholar * Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM . (2007). Accuracy and quality
of massively parallel DNA pyrosequencing. _Genome Biol_ 8: R143. Article PubMed PubMed Central Google Scholar * Huson DH, Auch AF, Qi J, Schuster SC . (2007). MEGAN analysis of
metagenomic data. _Genome Res_ 17: 377–386. Article CAS PubMed PubMed Central Google Scholar * Ikner LA, Toomey RS, Nolan G, Neilson JW, Pryor BM, Maier RM . (2007). Culturable
microbial diversity and the impact of tourism in Kartchner Caverns, Arizona. _Microb Ecol_ 53: 30–42. Article PubMed Google Scholar * Jagnow DH . (1999). Geology of Kartchner Caverns
State Park, Arizona. _J Cave Karst Stud_ 61: 49–58. Google Scholar * Jones DS, Albrecht HL, Dawson KS, Schaperdoth I, Freeman KH, Pi Y _et al_ (2012). Community genomic analysis of an
extremely acidophilic sulfur-oxidizing biofilm. _ISME J_ 6: 158–170. Article CAS PubMed Google Scholar * Kim BK, Jung MY, Yu DS, Park SJ, Oh TK, Rhee SK _et al_ (2011). Genome sequence
of an ammonia-oxidizing soil archaeon, "_Candidatus Nitrosoarchaeum koreensis_" My1. _J Bacteriol_ 193: 5539–5540. Article CAS PubMed PubMed Central Google Scholar * Konneke
M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. _Nature_ 437: 543–546. Article PubMed Google
Scholar * Kraft B, Strous M, Tegetmeyer HE . (2011). Microbial nitrate respiration—genes, enzymes and environmental distribution. _J Biotechnol_ 155: 104–117. Article CAS PubMed Google
Scholar * Laiz L, Groth I, Gonzalez I, Saiz-Jimenez C . (1999). Microbiological study of the dripping waters in Altamira Cave (Santillana del Mar, Spain). _J Microbiol Meth_ 36: 129–138.
Article CAS Google Scholar * Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S _et al_ (2009). The genomic basis of trophic strategy in marine bacteria. _Proc Natl Acad Sci
USA_ 106: 15527–15533. Article CAS PubMed PubMed Central Google Scholar * Legatzki A, Ortiz M, Neilson JW, Casavant RR, Palmer MW, Rasmussen C _et al_ (2012). Factors influencing
observed variations in the structure of bacterial communities on calcite formations in Kartchner Caverns, AZ, USA. _Geomicrobiol J_ 29: 422–434. Article Google Scholar * Legatzki A, Ortiz
M, Neilson JW, Dominguez S, Andersen GL, Toomey RS _et al_ (2011). Bacterial and archaeal community structure of two adjacent calcite speleothems in Kartchner Caverns, Arizona, USA.
_Geomicrobiol J_ 28: 99–117. Article Google Scholar * Lucker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B _et al_ (2010). A _Nitrospira_ metagenome illuminates the physiology
and evolution of globally important nitrite-oxidizing bacteria. _Proc Natl Acad Sci USA_ 107: 13479–13484. Article PubMed PubMed Central Google Scholar * Macalady JL, Jones DS, Lyon EH .
(2007). Extremely acidic, pendulous cave wall biofilms from the Frasassi Cave system, Italy. _Environ Microbiol_ 9: 1402–1414. Article CAS PubMed Google Scholar * Markert S, Arndt C,
Felbeck H, Becher D, Sievert SM, Hugler M _et al_ (2007). Physiological proteomics of the uncultured endosymbiont of _Riftia pachyptila_. _Science_ 315: 247–250. Article CAS PubMed Google
Scholar * Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC . (2009). IMG ER: A system for microbial genome annotation expert review and curation. _Bioinformatics_ 25:
2271–2278. Article CAS PubMed Google Scholar * Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA . (2009). Ammonia oxidation kinetics determine niche separation of
nitrifying archaea and bacteria. _Nature_ 461: 976–979. Article CAS PubMed Google Scholar * Nakagawa S, Takai K . (2008). Deep-sea vent chemoautotrophs: diversity, biochemistry and
ecological significance. _FEMS Microbiol Ecol_ 65: 1–14. Article CAS PubMed Google Scholar * Norris PR, Davis-Belmar CS, Brown CF, Calvo-Bado LA . (2011). Autotrophic, sulfur-oxidizing
actinobacteria in acidic environments. _Extremophiles_ 15: 155–163. Article CAS PubMed Google Scholar * Northup DE, Barns SM, Yu LE, Spilde MN, Schelble RT, Dano KE _et al_ (2003).
Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. _Environ Microbiol_ 5: 1071–1086. Article PubMed Google Scholar * Ortiz M, Neilson JW,
Nelson WM, Legatzki A, Byrne A, Yu Y _et al_ (2013). Profiling bacterial diversity and taxonomic composition on speleothem surfaces in Kartchner Caverns, AZ. _Microb Ecol_ 65: 371–383.
Article PubMed Google Scholar * Pester M, Schleper C, Wagner M . (2011). The _Thaumarchaeota_: An emerging view of their phylogeny and ecophysiology. _Curr Opin Microbiol_ 14: 300–306.
Article CAS PubMed PubMed Central Google Scholar * Porca E, Jurado V, Zgur-Bertok D, Saiz-Jimenez C, Pasic L . (2012). Comparative analysis of yellow microbial communities growing on
the walls of geographically distinct caves indicates a common core of microorganisms involved in their formation. _FEMS Microbiol Ecol_ 81: 255–266. Article CAS PubMed Google Scholar *
Portillo MC, Gonzalez JM, Saiz-Jimenez C . (2008). Metabolically active microbial communities of yellow and grey colonizations on the walls of Altamira Cave, Spain. _J Appl Microbiol_ 104:
681–691. Article CAS PubMed Google Scholar * Pratscher J, Dumont MG, Conrad R . (2011). Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. _Proc
Natl Acad Sci USA_ 108: 4170–4175. Article CAS PubMed PubMed Central Google Scholar * Ramos-Vera WH, Berg IA, Fuchs G . (2009). Autotrophic carbon dioxide assimilation in
_Thermoproteales_ revisited. _J Bacteriol_ 191: 4286–4297. Article CAS PubMed PubMed Central Google Scholar * Rütting T, Boeckx P, Müller C, Klemedtsson L . (2011). Assessment of the
importance of dissimilatory nitrate reduction to ammonium for the terrestrial nitrogen cycle. _Biogeosciences_ 8: 1779–1791. Article Google Scholar * Sarbu SM, Kane TC, Kinkle BK . (1996).
A chemoautotrophically based cave ecosystem. _Science_ 272: 1953–1955. Article CAS PubMed Google Scholar * Simon KS, Benfield EF, Macko SA . (2003). Food web structure and the role of
epilithic biofilms in cave streams. _Ecology_ 84: 2395–2406. Article Google Scholar * Solis-Dominguez FA, Valentin-Vargas A, Chorover J, Maier RM . (2011). Effect of arbuscular mycorrhizal
fungi on plant biomass and the rhizosphere microbial community structure of mesquite grown in acidic lead/zinc mine tailings. _Sci Total Environ_ 409: 1009–1016. Article CAS PubMed
PubMed Central Google Scholar * Spear JR, Barton HA, Robertson CE, Francis CA, Pace NR . (2007). Microbial community biofabrics in a geothermal mine adit. _Appl Environ Microbiol_ 73:
6172–6180. Article CAS PubMed PubMed Central Google Scholar * Strauss G, Eisenreich W, Bacher A, Fuchs G . (1992). 13c-nmr study of autotrophic CO2 fixation pathways in the
sulfur-reducing archaebacterium _Thermoproteus neutrophilus_ and in the phototrophic eubacterium _Chloroflexus aurantiacus_. _Eur J Biochem_ 205: 853–866. Article CAS PubMed Google
Scholar * Tetu SG, Breakwell K, Elbourne LD, Holmes AJ, Gillings MR, Paulsen IT . (2013). Life in the dark: Metagenomic evidence that a microbial slime community is driven by inorganic
nitrogen metabolism. _ISME J_ 7: 1227–1236. Article CAS PubMed PubMed Central Google Scholar * Toomey RS, Nolan G (2005). Environmental change at Kartchner Caverns: Trying to separate
natural and anthropogenic changes. In: Gottfried GJ, Gebow BS, Eskew LG, Edminster CB (eds.) _Connecting mountain islands and desert seas: Biodiveristy and management of the Madrean
ArchipelagoII. Proceedings RMRS-P-36, US Department of Agriculture, Forest Service, Rocky Mountain Research Station: Fort Collins, CO._ pp 264–270. * Tufts R, Tenen G . (1999). Discovery and
history of Kartchner Caverns, Arizona. _J Cave Karst Stud_ 61: 44–48. Google Scholar * Utaker JB, Andersen K, Aakra A, Moen B, Nes IF . (2002). Phylogeny and functional expression of
ribulose 1,5-bisphosphate carboxylase/oxygenase from the autotrophic ammonia-oxidizing bacterium _Nitrosospira sp_. Isolate 40ki. _J Bacteriol_ 184: 468–478. Article CAS PubMed PubMed
Central Google Scholar * Wang Y, Cheng H, Edwards RL, He Y, Kong X, An Z _et al_ (2005). The holocene asian monsoon: Links to solar changes and north atlantic climate. _Science_ 308:
854–857. Article CAS PubMed Google Scholar * Zhang LM, Offre PR, He JZ, Verhamme DT, Nicol GW, Prosser JI . (2010). Autotrophic ammonia oxidation by soil thaumarchaea. _Proc Natl Acad
Sci USA_ 107: 17240–17245. Article CAS PubMed PubMed Central Google Scholar * Zhou J, Gu Y, Zou C, Mo M . (2007). Phylogenetic diversity of bacteria in an earth-cave in Guizhou
Province, Southwest of China. _J Microbiol_ 45: 105–112. CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We express our appreciation to Robert Casavant, Ginger Nolan and
Steve Willsey of Arizona State Parks for their assistance in Kartchner Caverns and Nick Sisneros, and Yeisoo Yu of the Arizona Genomics Institute for extensive assistance with
pyrosequencing. Funding for this work was provided by the National Science Foundation Microbial Observatory grant MCB0604300 and a University of Arizona National Science Foundation IGERT
Genomics Initiative fellowship awarded to Marianyoly Ortiz, grant no. DGE-0654423. This work represents an original research paper. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department
of Soil, Water and Environmental Science, University of Arizona, Tucson, AZ, USA Marianyoly Ortiz, Antje Legatzki, Julia W Neilson & Raina M Maier * Department of Computer Science,
University of Arizona, Tucson, AZ, USA Brandon Fryslie * BIO5 Institute, Tucson, AZ, USA William M Nelson & Carol A Soderlund * Department of Plant Sciences, University of Arizona,
Tucson, AZ, USA Rod A Wing & Barry M Pryor Authors * Marianyoly Ortiz View author publications You can also search for this author inPubMed Google Scholar * Antje Legatzki View author
publications You can also search for this author inPubMed Google Scholar * Julia W Neilson View author publications You can also search for this author inPubMed Google Scholar * Brandon
Fryslie View author publications You can also search for this author inPubMed Google Scholar * William M Nelson View author publications You can also search for this author inPubMed Google
Scholar * Rod A Wing View author publications You can also search for this author inPubMed Google Scholar * Carol A Soderlund View author publications You can also search for this author
inPubMed Google Scholar * Barry M Pryor View author publications You can also search for this author inPubMed Google Scholar * Raina M Maier View author publications You can also search for
this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Julia W Neilson. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL
INFORMATION Supplementary Information accompanies this paper on The ISME Journal website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (DOC 362 KB) RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Ortiz, M., Legatzki, A., Neilson, J. _et al._ Making a living while starving in the dark: metagenomic insights into the energy dynamics of a
carbonate cave. _ISME J_ 8, 478–491 (2014). https://doi.org/10.1038/ismej.2013.159 Download citation * Received: 18 April 2013 * Revised: 03 August 2013 * Accepted: 12 August 2013 *
Published: 12 September 2013 * Issue Date: February 2014 * DOI: https://doi.org/10.1038/ismej.2013.159 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
KEYWORDS * chemoautotrophy * comparative metagenomics * archaea * carbonate cave * oligotrophy * aphotic habitat
