Fitness costs restrict niche expansion by generalist niche-constructing pathogens
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT We investigated the molecular and ecological mechanisms involved in niche expansion, or generalism, versus specialization in sympatric plant pathogens. Nopaline-type and
octopine-type _Agrobacterium tumefaciens_ engineer distinct niches in their plant hosts that provide different nutrients: nopaline or octopine, respectively. Previous studies revealed that
nopaline-type pathogens may expand their niche to also assimilate octopine in the presence of nopaline, but consequences of this phenomenon on pathogen dynamics in planta were not known.
Here, we provided molecular insight into how the transport protein NocT can bind octopine as well as nopaline, contributing to niche expansion. We further showed that despite the ability for
niche expansion, nopaline-type pathogens had no competitive advantage over octopine-type pathogens in co-infected plants. We also demonstrated that a single nucleotide polymorphism in the
_nocR_ gene was sufficient to allow octopine assimilation by nopaline-type strains even in absence of nopaline. The evolved _nocR_ bacteria had higher fitness than their ancestor in
octopine-rich transgenic plants but lower fitness in tumors induced by octopine-type pathogens. Overall, this work elucidates the specialization of _A. tumefaciens_ to particular opine
niches and explains why generalists do not always spread despite the advantage associated with broader nutritional niches. SIMILAR CONTENT BEING VIEWED BY OTHERS RAPID EVOLUTION OF BACTERIAL
MUTUALISM IN THE PLANT RHIZOSPHERE Article Open access 22 June 2021 EXTRACELLULAR NICHE ESTABLISHMENT BY PLANT PATHOGENS Article 08 January 2024 EVOLUTION OF RHIZOBIAL SYMBIOSIS ISLANDS
THROUGH INSERTION SEQUENCE-MEDIATED DELETION AND DUPLICATION Article Open access 16 July 2021 INTRODUCTION Niche construction designates the process by which a living population modifies its
environment and takes advantage of the induced environmental changes (Kylafis and Loreau, 2011). Niche construction processes help explain how sympatric populations can reduce direct
competition when coexisting in the same habitat, when each is able to construct and retrieve benefits from its particular niche. However, the resulting equilibrium might be threatened if a
population can also exploit niches that are constructed by others, a feature referred to as niche expansion that corresponds to an evolution towards a more generalist ecological behavior.
Indeed generalist strategies appear to have an advantage in fine-grained environments where individuals may encounter different habitat types or nutritional niches. The existence of several
specialists instead of a single generalist is therefore an evolutionary and ecological puzzle that remains unsolved despite long and active debates (Futuyama and Moreno, 1988; Palaima,
2007). All models that explore the conditions for the evolution and coexistence of specialists and generalists (for example, Wilson and Yoshimura, 1994; Egas et al., 2004; Abrams, 2006,
2012; Nurmi and Parvinen, 2008) assume that generalists trade off their ability to exploit multiple niches for their efficiency in any one. Indeed, were there no cost to being a generalist,
generalists would always replace specialists, so the coexistence of these two strategies requires the existence of such costs, which are, however, seldom evaluated (Kassen, 2002; Palaima,
2007; Satterwhite and Cooper, 2015; Schick et al., 2015). Here, work with the plant pathogen _Agrobacterium tumefaciens_ permits us to contribute new observations that help elucidate these
questions of the evolution of generalist versus specialist strategies and competition and coexistence of the two types of strategies within their constructed niches. The plant pathogen
_Agrobacterium tumefaciens_ is remarkable in its ability to generate specific nutrient niches in its host plants. On infection, _A. tumefaciens_ transfers a DNA-fragment (namely the T-DNA)
from the tumor-inducing (Ti) plasmid into the plant host cells (Pitzschke and Hirt, 2010). Once integrated into the plant host nuclear genome, the T-DNA genes drive niche construction by
producing a tumor within which low molecular weight (200–600 g mol−1) compounds, called opines, are synthesized. Only _A. tumefaciens_ cells that contain a Ti-plasmid can catabolize opines,
which are thus key players in _Agrobacterium_ niche construction (Guyon et al., 1980; Tempé and Petit, 1983). Indeed, bacterial cells harboring Ti-plasmids that catabolize opines outcompete
cells with mutant Ti-plasmids lacking opine catabolic activity, formally demonstrating the opine-niche concept (Lang et al., 2014). Importantly, as different Ti-plasmids code and catabolize
opines with a range of molecular structures (over twenty different opines described; (Dessaux et al., 1998), there may be several distinct opine niches in plant tumors, allowing the
coexistence of different _A. tumefaciens_ populations (Tempé and Petit, 1983; Bouzar and Moore, 1987). Nopaline and octopine are among the most extensively studied opines: nopaline is a
condensate of arginine and α-ketoglutarate, whereas octopine is a condensate of arginine and pyruvate (Dessaux et al., 1992, 1998). The nopaline-type _A. tumefaciens_ strains construct a
nopaline niche and assimilate nopaline, whereas the octopine-type _A. tumefaciens_ strains construct an octopine niche and assimilate octopine. Thus, these two pathogen populations should be
able to co-exist in distinct niches in the same plant tumor. However, early works revealed that the nopaline assimilative pathway also permits the importation and degradation of octopine
(Klapwijk et al., 1977; Zanker et al., 1992, 1994). Hence, nopaline-type strains are able to assimilate octopine once nopaline induces the nopaline assimilative pathway, giving them the
ability to expand their nutritional niche, potentially competing with octopine-strains and changing the conditions for stable coexistence. Combining structural biology, molecular microbial
ecology and plant genetics, we explored _A. tumefaciens_ niche construction and exploitation, testing for niche expansion and competition. Our results help explain how different _A.
tumefaciens_ opine types can persistently co-exist in the same host plant and even in the same tumor, despite the apparent ability of some _A. tumefaciens_ pathogens to exploit more than one
opine-niche. MATERIALS AND METHODS SYNTHESIS, DETECTION AND QUANTIFICATION OF OPINES Nopaline was synthetically obtained by condensation between l-arginine and α-ketoglutarate in the
presence of sodium cyanoborohydride as described by Tempé (1983). Octopine was synthetically obtained by condensation between l-arginine and l-2-bromopropionic acid (Tempé, 1983). The
detection and quantification of opines in whole extracts of plant tumors was done following a separation of compounds by high-voltage paper electrophoresis and chemical revelation using the
phenanthrene quinone reagent (Dessaux et al., 1992). PURIFICATION, CRYSTALLIZATION AND STRUCTURE DETERMINATION OF NOCT AND ITS M117N MUTANT IN COMPLEX WITH OCTOPINE The NocT protein of the
nopaline-type _A. tumefaciens_ C58 and NocT-M117N mutant were purified and co-crystallized with octopine instead of nopaline as previously described (Lang et al., 2014). High resolution
diffraction data were collected at 100 K on the PROXIMA I beamline at SOLEIL synchrotron (Saint-Aubin, France). Data collection and processing statistics are given in Supplementary Table S1.
Because the crystals of NocT and M117N mutant with octopine are isomorphous to those with nopaline, phase determination was straightforward. The resulting electron density maps showed the
presence of an octopine in the ligand binding site. Refinement was performed with BUSTER-2.10 (Blanc et al., 2004) with non crystallographic symmetry restraints as all asymmetric units
contain two protein molecules. One torsion libration screw-motion group was assigned for each structure. Electron density maps were evaluated using COOT (Emsley and Cowtan, 2004). Refinement
details are shown in Supplementary Table S1. Molecular graphics images were generated using PYMOL software (http://www.pymol.org). _K_D MEASUREMENTS BY FLUORESCENCE TITRATION AND
MICROCALORIMETRY Octopine bound to NocT was monitored by autofluorescence by exciting the protein at a wavelength of 295 nm and monitoring the quenching of fluorescence emission of
tryptophan residues at 335 nm. The experiment was performed at 22 °C in 3 × 15 mm quartz cuvettes using a Cary Eclypse spectrofluorometer (Varian, AgilentTechnologies, Santa Clara, CA, USA),
in 25 mm Tris–HCl pH 8 and 150 mm NaCl with a fixed amount of proteins (2 μm) and increasing concentrations of octopine. Each ligand had no emission signal at 335 nm. The data were analysed
using Origin 7 software (OriginLab Corp., Northampton, MA, USA) and fitted to the equation _f_=ΔFluorescencemax*abs(x)/(_K_D+abs(x)). Isothermal titration microcalorimetry experiments were
performed with an ITC200 isothermal titration calorimeter from MicroCal (GE Healthcare, Uppsala, Sweden). The experiments were carried out at 20 °C. Protein concentration in the
microcalorimeter cell (0.2 ml) was 100 μm. Nineteen injections of 2 μl of octopine solution at 1 mM were performed at 180 s intervals while stirring at 1000 r.p.m. The experimental data were
fitted to theoretical titration curves with the software supplied by MicroCal (ORIGIN). This software uses the relationship between the heat generated by each injection and Δ_H_ (enthalpy
change in Kcal Mol−1), Ka (the association binding constant in m−1), _n_ (the number of binding sites), total protein concentration and free and total ligand concentrations. BACTERIAL
STRAINS AND GROWTH CONDITIONS The octopine-type _A. tumefaciens_ strain R10 and nopaline-type _A. tumefaciens_ strain C58 were from our laboratory collection. _A. tumefaciens_ C58
derivatives carrying mutations in the Ti-plasmid had been previously constructed. They were: (i) C58 (pTi::Gm), that carries a gentamycin (Gm) cassette inserted into the _atu6147_ gene,
which is not involved in virulence, fitness in plant tumor or opine pathways (Haudecoeur et al., 2009; Lang et al., 2013); (ii) C58 (pTi-nos::Km) that carries a kanamycin (Km) cassette
inserted into the _nos_ gene (=_atu6015_), which is involved in the T-DNA-directed, nopaline synthesis in host plant; and, (iii) C58 (pTi-ocd::Gm) that contains a Gm resistance cassette
inserted into the _ocd_ gene (=_atu6016_), which is involved in nopaline catabolism (Lang et al., 2014). We imposed artificial selection for modified opine assimilation as follows: to select
for octopine assimilation ability in nopaline-type strains, overnight liquid cultures of nopaline-type _A. tumefaciens_ C58 (pTi::Gm) and C58 (pTi-nos::Km) were washed three times, adjusted
to OD600nm=1 and 3 ml of these cultures were plated onto agar AB medium (Chilton et al., 1974) containing 3 mm of octopine as sole source of carbon and nitrogen and incubated for 5 days.
Similarly, the octopine-type _A. tumefaciens_ R10 was cultivated with nopaline as a sole nutrient source. Characteristics of the _A. tumefaciens_ strains used in this study are summarized in
the Table 1. _A_ _. tumefaciens_ was cultivated at 30 °C in _Agrobacterium_ broth (AB) minimal medium supplemented with ammonium chloride (1 g l−1) and mannitol (2 g l−1) except when an
alternative source of carbon and nitrogen is indicated, or in Luria–Bertani modified medium (LBm, with 5 g l−1 NaCl). The antibiotics gentamycin and kanamycin were added at 25 μg ml−1 and
100 μg ml−1, respectively. DNA EXTRACTION, GENOME SEQUENCING AND VARIANT ANALYSIS DNA was extracted and purified with the DNeasy Blood and Tissue kit (Qiagen, Les Ulis, France) according to
the manufacturer’s instructions. For each clone, paired-end libraries (2 × 100) were prepared from 5 μg of total genomic DNA using the TruSeq SBS Kit v3—HS 200-cycles (FC-401-3001, Illumina,
Paris, France). Hiseq sequencing was performed at the Imagif platform (Gif-sur-Yvette, France) and the data were analyzed through the CASAVA-1.8.2 (Illumina; demultiplexing), Fastqc 0.10.1
(Babraham, UK; read quality) and Cutadapt-1.3 (Wilmington, DE, USA; adapter trimming) pipeline. Sequence reads were mapped on the annotated reference genome of _A. tumefaciens_ strain C58
(Wood et al., 2001). Mappings were carried out using the CLC Genomics Workbench v7.5 (CLC bio, Aarhus, Denmark) with a read length #0.9 and similarity #0.95. Genomic variant detection was
processed using CLC Genomics Workbench with a minimum coverage of 10 and a variant probability of 90%. RNA EXTRACTION AND GENE EXPRESSION ANALYSIS Total bacterial RNA was extracted from
early exponential phase cells using phenol-based protocol as previously described (Lang et al., 2013). cDNA was prepared from 1 μg of RNA using RevertAid H Minus First Strand cDNA Synthesis
Kit (Fermentas, Villebon, France) following the manufacturer’s instructions. Quantitative reverse transcription polymerase chain reactions (RT-qPCRs) were performed in a Lightcycler 480 II
(Roche, Meylan, France) apparatus. The data were processed using the 2-ΔΔCT method. For all primer sets the similarities of amplification efficiencies were controlled. The internal control
used was the gene _atu2422_. PLANT LINES, PLANT INFECTION AND QUANTIFYING BACTERIAL POPULATIONS All _Arabidopsis thaliana_ plants in this study are from the Columbia ecotype. The OCT+ plant
line that accumulates octopine was obtained by inserting the _A. tumefaciens_ octopine synthase gene under the control of a constitutive promoter into the _A. thaliana_ genome as previously
described (Mondy et al., 2014). Plants were grown in a greenhouse under long day conditions and controlled temperature (24–26 °C) for three weeks and then transferred into controlled
environment chambers (22 °C, 8 h photoperiod, 65% hygrometry). Young primary stems (about one-month old plants) were wounded with a needle and inoculated with agrobacteria as previously
described (Lang et al., 2013), which yielded a single tumor per plant. Crushed tumors at 32 days post-infection were suspended into 0.8% NaCl to recover the bacteria. Dilution series were
prepared and spread onto selective agar media for enumerating colony forming units (CFU). Selection was based either on antibiotic resistance or on ability to use nopaline/octopine as sole
sources of C and N. For mixed inoculations, an inoculum mix, consisting in equal proportions of the two or three strains, was plated onto non-selective medium. The ratios of the different
strains were estimated according to their distinctive phenotype and genotype. The proportions of the different bacterial genotypes were estimated by pooling counts from three independent
samples of each mixed inoculum (Pi) and from a sample of bacteria from each plant tumor (Pt). This allowed calculation of the competitive index CI=(Ptmutant/Ptcontrol)/(Pimutant/Picontrol)
as previously described (Macho et al., 2010). ACCESSION CODES Coordinates and structure factors were deposited at the Protein Data Bank (PDB) under accession code 5ITP for NocT-octopine and
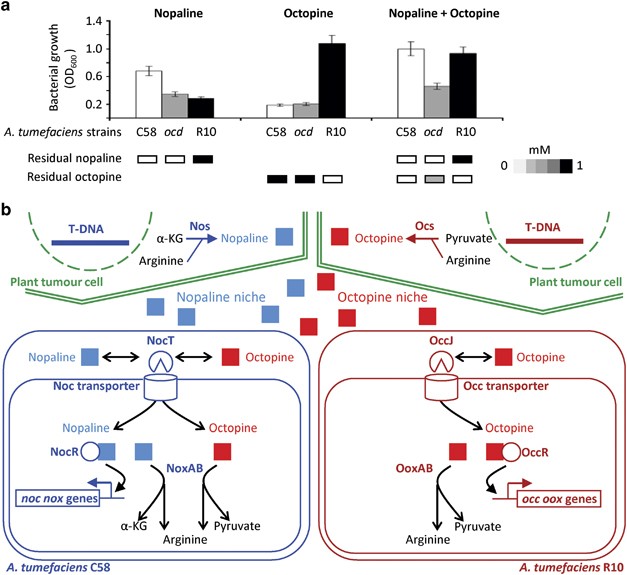
5ITO for NocT-M117N-octopine. RESULTS THE NOPALINE-TYPE PATHOGEN ASSIMILATED OCTOPINE IN THE PRESENCE OF NOPALINE The two pathogens _A. tumefaciens_ C58 (nopaline-type) and R10
(octopine-type) assimilated _in vitro_ the opines whose synthesis they induce in the host plants (Figure 1a). Furthermore, we observed that the nopaline-type strain could also metabolize
octopine in the presence of nopaline (Figure 1a). Using the _A. tumefaciens_ C58 _ocd_ mutant whose nopaline catabolism is reduced because its arginine assimilation is impaired (Sans et al.,
1987; Lang et al., 2014), we were able to infer the involvement of the nopaline catabolic genes in the growth capacity conferred by octopine (Figure 1a). From these data and previous
studies (Klapwijk et al., 1977; Zanker et al., 1992), it appeared that _A. tumefaciens_ C58 assimilates octopine via the nopaline catabolism pathway, the expression of which is only
activated by nopaline. The main molecular actors of opine-niche construction and utilization by the nopaline-type and octopine-type _A. tumefaciens_ pathogens are presented in Figure 1b. THE
OCTOPINE- AND NOPALINE-TYPE _A. TUMEFACIENS_ CO-EXISTED IN THE PLANT TUMOR Wild-type _A. thaliana_ plants were infected with a mixed inoculum of approximately 1:1 nopaline-type _A.
tumefaciens_ C58 and octopine-type _A. tumefaciens_ R10. The plant tumor niches were analyzed 32 days post-infection. Opine quantification revealed that both nopaline (200 pmol mg−1 fresh
plant tissues) and octopine (50 pmol mg−1 fresh plant tissues) accumulated in each plant tumor. In these same plant tumors, the total _A. tumefaciens_ population reached a mean (±s.d.) of
2.07 × 106±1.54 × 106 CFU mg−1 fresh plant tissues, and the strain ratio did not differ significantly from that of the inoculum (Fisher’s exact test, _P_=0.08). This indicated that the two
_A. tumefaciens_ opine types, which constructed two different opine niches, co-existed in each plant tumor. Furthermore, though _A. tumefaciens_ C58 can also assimilate octopine in the
presence of nopaline, it did not outcompete _A. tumefaciens_ strain R10, which only assimilates octopine. In fact, the proportion of the generalist C58 had somewhat, albeit
non-significantly, decreased from inoculum to tumors. Thus the generalist phenotype, able to assimilate and catabolize both nopaline and octopine, appeared to have no advantage over the
specialist octopine-type that benefits from a more restrictive nutritional niche under these conditions. THE IMPORT SYSTEM OF NOPALINE ACCOMMODATES BOTH NOPALINE AND OCTOPINE The periplasmic
nopaline-binding protein NocT is an essential molecular actor in the recognition and uptake of nopaline (Lang et al., 2014; Figure 1b). We investigated, at the atomic level, the binding
mode and the specificity of NocT towards nopaline and octopine. We solved the X-ray structure of the NocT in complex with octopine at 1.85 Å resolution. Crystals of NocT in complex with
nopaline or octopine are isomorphous and contained two very similar molecules in the asymmetric unit. All liganded NocT structures with nopaline and octopine are also very similar with an
overall root mean square deviation for all Cα atoms between 0.2 Å and 0.4 Å. The octopine located at the interface between the two closed lobes are very well defined in its electron density
maps (Figure 2a). On superposition of octopine and nopaline NocT complexes, both ligands overlap making identical protein interactions for their common arginine part (Figure 2b).
Interestingly, arginine alone cannot bind to NocT (Lang et al., 2014). The presence of α-ketoglurate (α-KG) for nopaline and pyruvate for octopine is thus required for protein binding. The
ligand binding site is composed of eight hydrophobic residues including Met117 which stabilized the pyruvate moiety of octopine by Van der Walls contacts, as previously observed for the α-KG
in nopaline (Lang et al., 2014). In addition, we solved the structure of the NocT-M117N mutant with octopine at 2.35 Å resolution. This NocT mutant was previously designed to demonstrate
the key-role of Met117 in nopaline affinity (Lang et al., 2014). The structure of NocT-M117N in complex with octopine resembles that of wild-type NocT in complex with octopine except that
the asparagine makes less contact with the ligand than the methionine (Supplementary Figure S1). THE OPINE-BINDING PROTEIN NOCT DISPLAYED A SIMILAR AFFINITY FOR OCTOPINE AND NOPALINE
Fluorescence titration experiments yielded an apparent _K_D value in the micromolar range with 6.1±0.7 μm for NocT towards octopine (Figure 3), in the same order of magnitude than that for
nopaline, 3.7 μm (Lang et al., 2014). Moreover, the isothermal titration microcalorimetry data confirmed the 1:1 binding stoichiometry with a mean KD of 9.9±0.7 μm (Figure 3). The octopine
binding in NocT presented a positive enthalpy change suggesting that its binding mode is entropy driven. Remarkably, the M117N mutant displayed a _K_D of 68.9±3.9 μm for octopine that was
seven- and tenfold higher (according to microcalorimetry and autofluorescence titrations, respectively) than that of the wild-type protein. This showed that the replacement of Met117 with
Asn led to a loss of hydrophobic interactions with the ligand octopine. These affinity measurements demonstrated that the opine-trapping protein NocT does not discriminate between nopaline
and octopine. EMERGENCE OF GENERALIST USERS OF NOPALINE AND OCTOPINE FROM _A. TUMEFACIENS_ C58 Nopaline-type _A. tumefaciens_ C58 requires nopaline to be able to assimilate octopine, and
this requirement could constrain exploitation of the octopine niche by a nopaline-type _A. tumefaciens_ if it colonizes a pre-formed octopine niche. We tested whether variants able to
assimilate octopine even in the absence of nopaline can arise under the appropriate environmental conditions. After a five-day culture on agar plates containing octopine as sole source of C
and N, we isolated several derivatives of _A. tumefaciens_ C58 that were able to grow on octopine even in the absence of nopaline (Figure 4a). This observation indicated the presence of
genetic variation in nopaline-type _A. tumefaciens_ C58 for the ability to exploit a broader niche, without induction by nopaline. This nopaline-type _A. tumefaciens_ strain could not only
assimilate octopine in addition to its specific nopaline niche, it was also able to evolve the ability to assimilate octopine in a constitutive manner. Noticeably, the reciprocal experiment
in which _A. tumefaciens_ R10 cells were plated onto a medium containing only nopaline as source of C and N yielded no nopaline-user clones, suggesting that niche expansion onto nopaline is
less likely in this strain. This observation is in agreement with previous data showing that the catabolic complex NoxAB from nopaline-type bacteria can cleave both nopaline and octopine,
whereas the enzymatic complex OoxAB degrades octopine only, hence limiting the possibility of a niche expansion in octopine-type bacteria (Zanker et al., 1994). GENOME SCAN REVEALED THE
GENETIC TRAITS OF DERIVED OCTOPINE-USERS We isolated 20 clones able to catabolize octopine in the absence of nopaline, 11 (oct+1 to oct+11) derived from _A. tumefaciens_ C58 (pTi::Gm) and
nine derived from _A. tumefaciens_ C58 (pTi-nos::Km), which is unable to construct a nopaline niche in the plant host while retaining the capacity to import and assimilate both nopaline and
octopine (Table 1). These latter nine clones permitted us to carry out in planta assays without the complication of nopaline induction and possible interpretative ambiguities resulting from
nopaline and octopine interference. Growth assay confirmed that all of the derived clones could use both octopine and nopaline as sole source of C and N, unlike their ancestral strain
(Figure 4a for a subset). The genomes of 11 of these, five derived from _A. tumefaciens_ C58 (pTi::Gm) and six from _A. tumefaciens_ C58 (pTi-nos::Km), were sequenced. Each derived strain
presented between 1 and 4 nucleotide changes compared with their ancestral strain (Supplementary Table S2). Strikingly, all eleven clones displayed a nucleotide modification in the coding
sequence of the _nocR_ gene carried by the Ti-plasmid: eight at the position 40906 (T>C), one at the position 40915 (T>C), one at the position 40759 (G>A) and one at the position
41139 (C>T), all causing missense mutations (Figure 4a). NocR is a LysR-type transcriptional factor that controls, through binding with nopaline, the expression of the nopaline transport
(_noc_ operon) and catabolism (_nox_ operon) genes (von Lintig et al., 1991; Figure 1b). We evaluated the expression of the _nocT_ and _noxB_ genes in octopine-users exhibiting the different
variations in _nocR_ sequence, and, compared with their ancestral strains, all exhibited from a 50 and to almost a 1000-fold increase in expression even in the absence of opines (Figure
4b). DERIVED OCTOPINE-USERS ACQUIRED A SELECTIVE ADVANTAGE ONLY IN AN OCTOPINE-RICH PLANT ENVIRONMENT We investigated whether the octopine-users derived from nopaline-type _A. tumefaciens_
might be selected in the octopine niche of a plant tumor. To be sure that nopaline interfering with the expression of the nopaline/octopine assimilation pathway was not produced in plant
tumors, we used only _A. tumefaciens_ C58 (pTi-nos::Km) derivatives that are unable to induce the nopaline niche. The derived octopine-users oct+13 and oct+14 were co-infected with the
parental strain at approximately a 1:1 ratio in wild-type and in OCT+ _A. thaliana_ plants, genetically modified to synthetize and accumulate octopine (Mondy et al., 2014). In all assays,
the octopine contents and the proportion of competing strains were measured in plant tumors and the latter was compared with the strain proportion in the inoculum. In octopine- and
nopaline-free plant tumors induced on wild-type plants, both derived octopine-users oct+13 and oct+14 were disadvantaged in the presence of their parent (Figure 5a; Supplementary Figure S2),
with a mean (±s.d.) competitive index for oct+13 and oct+14 compared with their parent of only 0.33±0.17 and 0.25±0.18, respectively. In contrast, in the octopine-rich (about 800 pmol mg−1
of fresh plant tissue) plant environment provided by the _A. thaliana_ OCT+ line, the derived octopine-users oct+13 and oct+14 increased their representation compared with their parent
(Figure 5b), with a mean (±s.d.) competitive index of 2.96±1.88 and 3.92±2.81, respectively. Hence, derived octopine-users may be selected in an octopine-rich environment, but they were
disadvantaged in the absence of octopine and nopaline, that is, in an opine-poor environment. _DERIVED OCTOPINE-USERS WERE DISADVANTAGED IN_ A. TUMEFACIENS _R10-INDUCED OCTOPINE-TUMORS_ We
co-inoculated wild-type _A. thaliana_ plants with a triple mix (at a ratio of approximately 1:1:1) containing the _A. tumefaciens_ R10 strain that induces the octopine niche, _A.
tumefaciens_ C58 (pTi-nos::Km) and a derived octopine-user (oct+13). The tumors accumulated octopine (about 200 pmol mg−1 of fresh plant tissue) but no nopaline (Supplementary Figure S3).
Under these conditions, the proportion of the _A. tumefaciens_ R10 compared with the two _A. tumefaciens_ C58 strains together (derived plus ancestral strains) did not change significantly
between the inoculum and the plant tumor (Fisher’s exact test, _P_=0.7). This showed that a mixed _A. tumefaciens_ C58 population of nopaline-dependent and derived, octopine-users could not
outcompete _A. tumefaciens_ R10 that induces and uses octopine. In the same plant tumors, when considering just the relative proportion of the two C58 strains, the derived octopine-user
decreased in proportion compared with its parent between the inoculum and the tumor (Figure 5c). The competitive index of the derived strain was only 0.5. Taken together these results
established that a derived octopine-user that appears to arise easily from nopaline-type _A. tumefaciens_ C58, can gain a selective advantage in an artificially high-octopine environment
(Figure 5b but not in a more natural _A. tumefaciens_ R10-induced octopine niche (Figure 5c). These features may explain why we do not observe cases of the expanded niche arising by natural
selection in populations of nopaline-type pathogens. SIMULTANEOUS CONSTRUCTION OF TWO OPINE-TYPE NICHES AFFECTS THE OPINE RESOURCE The above data strongly suggested that the abundance of
octopine in the plant tumor is crucial in determining the fitness cost or benefit of octopine-users derived from nopaline-type pathogens. Therefore, we investigated factors affecting opine
abundance. We tested whether the simultaneous construction of two different opine-type niches in the same plant tumor could alter opine accumulation. We infected host plants with the
different opine-type pathogens alone and in combination, and measured their opine content at 32 days post-infection. Supplementary Figure S3 summarizes our opine-content data. The presence
of two infecting opine-type strains that induce two different opines dramatically decreased both octopine and nopaline levels compared with the tumors generated from infections of a single
opine-type. Using the _A. tumefaciens_ C58 (pTi-nos::Km) mutant that is defective in nopaline synthesis, we showed that the important effect arose from the induction of a second opine-niche,
and not the presence of a second bacterial strain. By modifying the production and accumulation of the opine resources, two concomitant niche constructions influenced the selection regime
experienced by the two coexisting pathogens. Though the generalist strategy generates an advantage under high resource levels, the low resource levels related to competing niche construction
prevented the spread of a generalist mutant pathogen, thereby maintaining specialist strategies in the _A. tumefaciens_ pathogen. DISCUSSION Octopine-type and nopaline-type populations of
_Agrobacterium tumefaciens_ may co-exist in the same host plant tumors, each pathogen population being able to construct a niche in which a distinctive opine accumulates and is used as a
nutrient. Surprisingly however, these two pathogen types are not equally specialized on their particular niche. Several traits allow nopaline-type pathogens to also exploit the octopine
niche: (i) the opine-trapping protein NocT of the nopaline-type pathogen _A. tumefaciens_ C58 recognized both nopaline and octopine, with a very close affinity, in the micromolar range,
meaning that NocT-mediated transport system had no preference between these opines (this work); (ii) the NoxAB enzyme complex, which releases arginine from nopaline and octopine, exhibits a
similar affinity and specific activity toward the two opines (Zanker et al., 1994); and (iii) when expressed, the nopaline/octopine-assimilative pathway conferred a growth advantage to
nopaline-type _A. tumefaciens_ in the presence of either nopaline alone or nopaline and octopine (Figure 1). Altogether, a nopaline-type _A. tumefaciens_ pathogen exhibits the transport and
metabolic traits required to exploit an octopine niche in a nopaline-dependent manner. However, despite a versatile metabolism towards opines, the more generalist nopaline-type pathogen
failed to outcompete the specialist octopine-type one in co-infected plant hosts, even showing a tendency to lower growth within the mixed plant tumors. Hence, hidden costs of this more
generalist strategy must outweigh the apparent advantages inherent in the broader niche. Such costs of a generalist strategy are implicitly assumed in numerous models (see Palaima, 2007 for
discussion) but have seldom been demonstrated (Kassen, 2002). Though we have not identified the precise nature of these costs or their mechanisms, our results suggest their existence in this
system, which explains why the generalist nopaline-octopine-user does not replace the octopine specialist. Alternatively, environmental conditions within the tumor, where octopine is
produced at lower concentrations than is nopaline, constrain the ability of the generalist to profit from this advantage. Indeed recent models exploring the conditions for
specialist-generalist coexistence showed generalists to suffer more from competition for a rare resource than do specialists (Abrams, 2012). Though living systems seldom replicate exactly
the idealized conditions of models, the _A. tumefaciens_ strains we used reflect quite well the conditions explored in Abrams (2012) models. Several constitutive octopine-users arose when
nopaline-type strains were cultivated on octopine as sole C and N source. The novel strains arose _via_ single nucleotide variations in the regulatory gene _nocR_, which permitted
nopaline-independent expression of the nopaline/octopine-assimilative pathway (Supplementary Table S2), similar to the observation of Akakura and Winans (2002). We tested whether this
constitutive ability to assimilate octopine could lead to a selective advantage for the derived nopaline-type _A. tumefaciens_. The constitutive variants of NocR indeed enjoyed a competitive
advantage in transgenic, octopine-rich host plants that accumulated large amounts of octopine. However, competition assays in the natural octopine niche that was induced by the
octopine-type pathogen revealed that the constitutive octopine-users deriving from the nopaline-type _A. tumefaciens_ did not outcompete their ancestor but exhibited indeed a reduced fitness
compared with the ancestral strain. This implies a cost associated with the new mutations that outweighed the advantage of niche expansion in the natural opine niche. Indeed, the mutant
strains all exhibited strong, constitutive overexpression of the noc operon, which could impose a physiological cost. As a consequence, even though constitutive octopine-users might arise by
_de novo_ mutation in progeny of nopaline-type _A. tumefaciens_, they are unlikely to spread in the induced plant opine niche. This implies that the acquisition of the new ability to
exploit octopine in a constitutive way, though opening new ecological opportunities for the invasion of octopine niches without needing the presence of nopaline to exploit them, imposed
immediate costs that constrain the evolution of the generalist strategy (Futuyama and Moreno, 1988). Another interesting result from our study is that tumors resulting from mixed infections
of two opine types, with simultaneous construction of the octopine and nopaline niches, accumulated less opine than did tumors resulting from single infections (Supplementary Figure S3). It
appears that the concomitant induction of the two opine niches affected one another. The mechanism by which this occurs remains unknown, but different parasite genotypes infecting the same
host may interact in a diversity of ways, including interference or cooperation (Buckling and Brockhurst, 2008). Nopaline and octopine both require arginine as precursor for their synthesis
(Figure 1b), so the two strains compete for this common substrate whose concentration in plants is tightly regulated (Winter et al., 2015). However this does not, in itself, explain why the
induction of a second opine-niche should reduce the overall production of opines, unless there is some direct interference. The observation, that the simultaneous induction of two opine
niches is less efficient than the induction of a single one, however, provides an additional constraint to the niche expansion process. We observed that the broader feeding niche containing
more than one induced opine provided a poorer environment with lower overall resource availability. As the derived generalist octopine exploiter had a competitive advantage only at
unnaturally high-octopine levels, the reduction in resource levels during co-infection will slow or prevent the spread of the generalist strategy (see also Abrams, 2012). Moreover the fact
that NocT had similar affinities for nopaline and octopine suggests that the most abundant opine nopaline could be the most frequently imported. This may explain why the ability to exploit
octopine in addition to nopaline granted no advantage under natural concentrations of these two opines. Altogether this work shows that niche expansion may be limited by several mechanisms,
thereby explaining the maintenance of multiple specialist strategies in place of a single, generalist one (Futuyama and Moreno, 1988) in _A. tumefaciens_ populations. Because other soil
Proteobacteria such as _Pseudomonas_ or _Ensifer_ (Wilson et al., 1995; Oger et al., 1997; Savka and Farrand, 1997; Oger et al., 2004; Mondy et al., 2014) may exploit the opine niches that
are constructed by the _Agrobacterium_ pathogens, the question of generalist versus specialist strategy remains to be explored in these niche opportunistic populations of the plant
microbiota. REFERENCES * Abrams PA . (2006). The prerequisites for and likelihood of generalist-specialist coexistence. _Am Nat_ 167: 329–342. Google Scholar * Abrams PA . (2012). The
eco-evolutionary responses of a generalist consumer to resource competition. _Evolution_ 66: 3130–3143. Article Google Scholar * Akakura R, Winans SC . (2002). Constitutive mutations of
the OccR regulatory protein affect DNA bending in response to metabolites released from plant tumors. _J Biol Chem_ 277: 5866–5874. Article CAS Google Scholar * Blanc E, Roversi P,
Vonrhein C, Flensburg C, Lea SM, Bricogne G . (2004). Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. _Acta Crystallogr D Biol Crystallogr_ 60: 2210–2221.
Article CAS Google Scholar * Bouzar H, Moore LW . (1987). Isolation of different agrobacterium biovars from a natural oak savanna and tallgrass prairie. _Appl Environ Microbiol_ 53:
717–721. CAS Google Scholar * Buckling A, Brockhurst MA . (2008). Kin selection and the evolution of virulence. _Heredity_ 100: 484–488. Article CAS Google Scholar * Chilton MD, Currier
TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW . (1974). _Agrobacterium tumefaciens_ DNA and PS8 bacteriophage DNA not detected in crown gall tumors. _Proc Natl Acad Sci USA_ 71:
3672–3676. Article CAS Google Scholar * Dessaux Y, Petit A, Tempe J . (1992). Opines in _Agrobacterium_ biology. In: Verma DPS (ed). _Molecular Signals In Plant–Microbe Communications_.
CRC Press: Boca Raton, FL, USA, pp 109–136. Google Scholar * Dessaux Y, Petit A, Farrand SK, Murphy PJ . (1998). Opines and opine-like molecules involved in Plant-Rhizobiaceae Interactions.
In: Spaink HP, et al. (eds). _The Rhizobiaceae, Molecular Biology of Model Plant-associated Bacteria_. Kluwer Academic Publisher: Dordrecht, The Netherlands, pp 173–197. Google Scholar *
Egas M, Dieckmann U, Sabelis MW . (2004). Evolution restricts the coexistence of specialists and generalists: The role of trade-off structure. _Am Nat_ 163: 518–531. Article Google Scholar
* Emsley P, Cowtan K . (2004). Coot: model-building tools for molecular graphics. _Acta Crystallogr D Biol Crystallogr_ 60: 2126–2132. Article Google Scholar * Futuyama DJ, Moreno G .
(1988). The evolution of ecological specialization. _Ann Rev Ecol Syst_ 19: 207–233. Article Google Scholar * Guyon P, Chilton MD, Petit A, Tempe J . (1980). Agropine in ‘null-type’ crown
gall tumors: Evidence for generality of the opine concept. _Proc Natl Acad Sci USA_ 77: 2693–2697. Article CAS Google Scholar * Haudecoeur E, Tannières M, Cirou A, Raffoux A, Dessaux Y,
Faure D . (2009). Different regulation and roles of lactonases AiiB and AttM in _Agrobacterium tumefaciens_ C58. _Mol Plant Microbe Interact_ 22: 529–537. Article CAS Google Scholar *
Kassen R . (2002). The experimental evolution of specialists, generalists, and the maintenance of diversity. _J Evol Biol_ 15: 173–190. Article Google Scholar * Klapwijk PM, Oudshoorn M,
Schilperoort RA . (1977). Inducible permease involved in the uptake of octopine, lysopine and octopinic acid by _Agrobacterium tumefaciens_ strains carrying virulence-associated plasmids.
_Microbiology_ 102: 1–11. CAS Google Scholar * Kylafis G, Loreau M . (2011). Niche construction in the light of niche theory. _Ecol Lett_ 14: 82–90. Article Google Scholar * Lang J,
Vigouroux A, Planamente S, El Sahili A, Blin P, Aumont-Nicaise M _et al_. (2014). _Agrobacterium_ uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage
in the niche construction on plant host. _PLoS Pathog_ 10: e1004444. Article Google Scholar * Lang J, Planamente S, Mondy S, Dessaux Y, Morera S, Faure D . (2013). Concerted transfer of
the virulence Ti plasmid and companion At plasmid in the _Agrobacterium tumefaciens_-induced plant tumour. _Mol Microbiol_ 90: 1178–1189. Article CAS Google Scholar * Macho AP, Guidot A,
Barberis P, Beuzon CR, Genin S . (2010). A competitive index assay identifies several _Ralstonia solanacearum_ type III effector mutant strains with reduced fitness in host plants. _Mol
Plant Microbe Interact_ 23: 1197–1205. Article CAS Google Scholar * Mondy S, Lenglet A, Beury-Cirou A, Libanga C, Ratet P, Faure D _et al_. (2014). An increasing opine carbon bias in
artificial exudation systems and genetically modified plant rhizospheres leads to an increasing reshaping of bacterial populations. _Mol Ecol_ 23: 4846–4861. Article Google Scholar * Nurmi
T, Parvinen K . (2008). On the evolution of specialization with a mechanistic underpinning in structured metapopulations. _Theor Pop Biol_ 73: 222–243. Article Google Scholar * Oger P,
Petit A, Dessaux Y . (1997). Genetically engineered plants producing opines alter their biological environment. _Nat Biotechnol_ 15: 369–372. Article CAS Google Scholar * Oger PM,
Mansouri H, Nesme X, Dessaux Y . (2004). Engineering root exudation of _Lotus_ toward the production of two novel carbon compounds leads to the selection of distinct microbial populations in
the rhizosphere. _Microb Ecol_ 47: 96–103. Article CAS Google Scholar * Palaima A . (2007). The fitness cost of generalization: present limitations and future possible solutions. _Biol J
Linn Soc_ 90: 583–590. Article Google Scholar * Pitzschke A, Hirt H . (2010). New insights into an old story: _Agrobacterium_-induced tumour formation in plants by plant transformation.
_Embo J_ 29: 1021–1032. Article CAS Google Scholar * Sans N, Schroder G, Schroder J . (1987). The Noc region of Ti plasmid C58 codes for arginase and ornithine cyclodeaminase. _Eur J
Biochem_ 167: 81–87. Article CAS Google Scholar * Satterwhite RS, Cooper TF . (2015). Constraints on adaptation of _Escherichia coli_ to mixed-resource environments increase over time.
_Evolution_ 69: 2067–2078. Article CAS Google Scholar * Savka MA, Farrand SK . (1997). Modification of rhizobacterial populations by engineering bacterium utilization of a novel
plant-produced resource. _Nat Biotechnol_ 15: 363–368. Article CAS Google Scholar * Schick A, Bailey SF, Kassen R . (2015). Evolution of fitness trade-offs in locally adapted populations
of _Pseudomonas fluorescens_. _Am Nat_ 186: S48–S59. Article Google Scholar * Tempé J, Petit A . (1983) La piste des opines. In: Pühler A (ed). _Molecular Genetics of the Bacteria–Plant
Interaction_. Springer-Verlag: Berlin-Heidelberg, pp 14–32. Book Google Scholar * Tempé J . (1983) Chemistry and biochemistry of open-chain imino-acids. In: Weistein B (ed). _Chemistry and
Biochemistry of Amino Acids, Peptides and Proteins_. Marcel Dekker Inc: New York, NY, USA, pp 113–203. Google Scholar * Von Lintig J, Zanker H, Schroder J . (1991). Positive regulators of
opine-inducible promoters in the nopaline and octopine catabolism regions of Ti plasmids. _Mol Plant Microbe Interact_ 4: 370–378. Article CAS Google Scholar * Wilson DS, Yoshimura J .
(1994). On the coexistence of specialists and generalists. _Am Nat_ 144: 692–707. Article Google Scholar * Wilson M, Savka MA, Hwang I, Farrand SK, Lindow SE . (1995). Altered epiphytic
colonization of mannityl opine-producing transgenic tobacco plants by a mannityl opine-catabolizing strain of _Pseudomonas syringae_. _Appl Environ Microbiol_ 61: 2151–2158. CAS Google
Scholar * Winter G, Todd CD, Trovato M, Forlani G, Funck D . (2015). Physiological implications of arginine metabolism in plants. _Front Plant Sci_ 6: 534. Article Google Scholar * Wood
DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK _et al_. (2001). The genome of the natural genetic engineer _Agrobacterium tumefaciens_ C58. _Science_ 294: 2317–2323. Article CAS
Google Scholar * Zanker H, Von Lintig J, Schroder J . (1992). Opine transport genes in the octopine (occ) and nopaline (noc) catabolic regions in Ti plasmids of _Agrobacterium tumefaciens_.
_J Bacteriol_ 174: 841–849. Article CAS Google Scholar * Zanker H, Lurz G, Langridge U, Langridge P, Kreusch D, Schroder J . (1994). Octopine and nopaline oxidases from Ti plasmids of
_Agrobacterium tumefaciens_: molecular analysis, relationship, and functional characterization. _J Bacteriol_ 176: 4511–4517. Article CAS Google Scholar Download references
ACKNOWLEDGEMENTS JL, AV, AK, SM and DF was supported by CNRS (Mission pour l’interdisciplinarité, Agromics 2014–2016) and ANR-Blanc SENSOR (ANR-12-BSV8-0003-01/02/03), and AES by a PhD-grant
of the University Paris-Saclay (ED 425). This work has benefited from the I2BC plant culture facilities, I2BC high-throughput sequencing platform and I2BC crystallization platform,
supported by FRISBI ANR-10-INSB-05-01. We acknowledge SOLEIL for provision of synchrotron radiation facilities (proposal ID 20130869) in using beamline Proxima I. AUTHOR INFORMATION Author
notes * Julien Lang Present address: Current address: IPS2, INRA, Gif-sur-Yvette 91 190, France., * Abbas El Sahili Present address: Current address: Division of Structural Biology &
Biochemistry | Nanyang Technological University Proteos, Singapore 138673 Singapore., * Anthony Kwasiborski Present address: Current address: Institut de Recherche en Horticulture et
Semences, UMR 1345 INRA—Université d'Angers-Agrocampus Ouest, 42 Rue Georges Morel—CS 60057, 49071 Beaucouzé Cedex, France., * Julien Lang and Armelle Vigouroux: These authors
contributed equally to this work. AUTHORS AND AFFILIATIONS * Institute for Integrative Biology of the Cell (I2BC), CNRS CEA Université Paris-Sud, Université Paris-Saclay, Gif-sur-Yvette,
France Julien Lang, Armelle Vigouroux, Abbas El Sahili, Anthony Kwasiborski, Magali Aumont-Nicaise, Yves Dessaux, Solange Moréra & Denis Faure * Ecologie Systématique Evolution, CNRS,
Université Paris-Sud, AgroParisTech, Université Paris-Saclay, Orsay, France Jacqui Anne Shykoff Authors * Julien Lang View author publications You can also search for this author inPubMed
Google Scholar * Armelle Vigouroux View author publications You can also search for this author inPubMed Google Scholar * Abbas El Sahili View author publications You can also search for
this author inPubMed Google Scholar * Anthony Kwasiborski View author publications You can also search for this author inPubMed Google Scholar * Magali Aumont-Nicaise View author
publications You can also search for this author inPubMed Google Scholar * Yves Dessaux View author publications You can also search for this author inPubMed Google Scholar * Jacqui Anne
Shykoff View author publications You can also search for this author inPubMed Google Scholar * Solange Moréra View author publications You can also search for this author inPubMed Google
Scholar * Denis Faure View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Solange Moréra or Denis Faure. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information accompanies this paper on The ISME Journal website
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (PDF 190 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Lang, J., Vigouroux, A., El Sahili, A.
_et al._ Fitness costs restrict niche expansion by generalist niche-constructing pathogens. _ISME J_ 11, 374–385 (2017). https://doi.org/10.1038/ismej.2016.137 Download citation * Received:
28 March 2016 * Revised: 04 August 2016 * Accepted: 07 September 2016 * Published: 01 November 2016 * Issue Date: February 2017 * DOI: https://doi.org/10.1038/ismej.2016.137 SHARE THIS
ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard
Provided by the Springer Nature SharedIt content-sharing initiative
