Visualizing virus assembly intermediates inside marine cyanobacteria
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

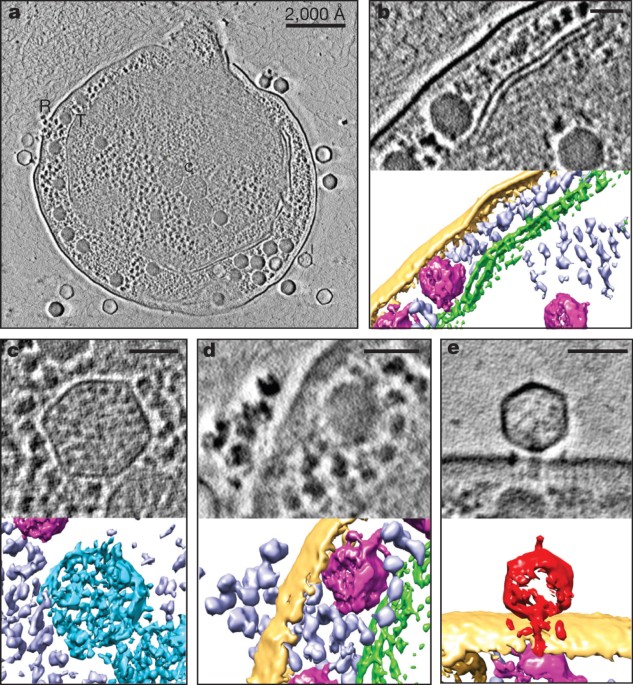
ABSTRACT Cyanobacteria are photosynthetic organisms responsible for ∼25% of organic carbon fixation on the Earth. These bacteria began to convert solar energy and carbon dioxide into
bioenergy and oxygen more than two billion years ago. Cyanophages, which infect these bacteria, have an important role in regulating the marine ecosystem by controlling cyanobacteria
community organization and mediating lateral gene transfer. Here we visualize the maturation process of cyanophage Syn5 inside its host cell, _Synechococcus_, using Zernike phase contrast
electron cryo-tomography (cryoET)1,2. This imaging modality yields dramatic enhancement of image contrast over conventional cryoET and thus facilitates the direct identification of
subcellular components, including thylakoid membranes, carboxysomes and polyribosomes, as well as phages, inside the congested cytosol of the infected cell. By correlating the structural
features and relative abundance of viral progeny within cells at different stages of infection, we identify distinct Syn5 assembly intermediates. Our results indicate that the procapsid
releases scaffolding proteins and expands its volume at an early stage of genome packaging. Later in the assembly process, we detected full particles with a tail either with or without an
additional horn. The morphogenetic pathway we describe here is highly conserved and was probably established long before that of double-stranded DNA viruses infecting more complex organisms.
Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this
journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now
Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS CRYO-EM STRUCTURE OF CYANOPHAGE P-SCSP1U OFFERS INSIGHTS INTO DNA GATING AND EVOLUTION OF T7-LIKE VIRUSES Article Open access 13 October 2023
FREQUENCY OF MISPACKAGING OF _PROCHLOROCOCCUS_ DNA BY CYANOPHAGE Article Open access 14 September 2020 STRUCTURE OF A THYLAKOID-ANCHORED CONTRACTILE INJECTION SYSTEM IN MULTICELLULAR
CYANOBACTERIA Article Open access 14 February 2022 ACCESSION CODES ACCESSIONS ELECTRON MICROSCOPY DATA BANK * EMD-5742 * EMD-5743 * EMD-5744 * EMD-5745 * EMD-5746 DATA DEPOSITS The averaged
density maps of the procapsid, expanded capsid and the DNA-containing capsid have been deposited in the EBI under accession codes EMD-5742, EMD-5743, EMD-5744, EMD-5745 and EMD-5746,
respectively. REFERENCES * Danev, R. & Nagayama, K. Phase plates for transmission electron microscopy. _Methods Enzymol._ 481, 343–369 (2010) Article Google Scholar * Murata, K. et al.
Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. _Structure_ 18, 903–912 (2010) Article CAS Google
Scholar * Fuller, N. J. et al. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine _Synechococcus_ clade throughout a stratified water column in the
red sea. _Appl. Environ. Microbiol._ 69, 2430–2443 (2003) Article CAS Google Scholar * Rocap, G., Distel, D. L., Waterbury, J. B. & Chisholm, S. W. Resolution of _Prochlorococcus_
and _Synechococcus_ ecotypes by using 16S–23S ribosomal DNA internal transcribed spacer sequences. _Appl. Environ. Microbiol._ 68, 1180–1191 (2002) Article CAS Google Scholar * Taylor, K.
A. & Glaeser, R. M. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. _J. Struct. Biol._ 163,
214–223 (2008) Article CAS Google Scholar * Danev, R., Glaeser, R. M. & Nagayama, K. Practical factors affecting the performance of a thin-film phase plate for transmission electron
microscopy. _Ultramicroscopy_ 109, 312–325 (2009) Article CAS Google Scholar * Marko, M., Leith, A., Hsieh, C. & Danev, R. Retrofit implementation of Zernike phase plate imaging for
cryo-TEM. _J. Struct. Biol._ 174, 400–412 (2011) Article CAS Google Scholar * Rochat, R. H. et al. Seeing the portal in herpes simplex virus type 1 B capsids. _J. Virol._ 85, 1871–1874
(2011) Article CAS Google Scholar * Liberton, M., Austin, J. R., II, Berg, R. H. & Pakrasi, H. B. Unique thylakoid membrane architecture of a unicellular N2-fixing cyanobacterium
revealed by electron tomography. _Plant Physiol._ 155, 1656–1666 (2011) Article CAS Google Scholar * Ting, C. S., Hsieh, C., Sundararaman, S., Mannella, C. & Marko, M. Cryo-electron
tomography reveals the comparative three-dimensional architecture of _Prochlorococcus_, a globally important marine cyanobacterium. _J. Bacteriol._ 189, 4485–4493 (2007) Article CAS Google
Scholar * Iancu, C. V. et al. Organization, structure, and assembly of α-carboxysomes determined by electron cryotomography of intact cells. _J. Mol. Biol._ 396, 105–117 (2010) Article
CAS Google Scholar * Schmid, M. F. et al. Structure of _Halothiobacillus neapolitanus_ carboxysomes by cryo-electron tomography. _J. Mol. Biol._ 364, 526–535 (2006) Article CAS Google
Scholar * Pope, W. H. et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine synechococcus. _J. Mol. Biol._ 368,
966–981 (2007) Article CAS Google Scholar * Chang, J. T. et al. Visualizing the structural changes of bacteriophage Epsilon15 and its _Salmonella_ host during infection. _J. Mol. Biol._
402, 731–740 (2010) Article CAS Google Scholar * Hu, B., Margolin, W., Molineux, I. J. & Liu, J. The bacteriophage t7 virion undergoes extensive structural remodeling during
infection. _Science_ 339, 576–579 (2013) Article ADS CAS Google Scholar * Schmid, M. F. Single-particle electron cryotomography (cryoET). _Adv. Protein Chem. Struct. Biol._ 82, 37–65
(2011) Article CAS Google Scholar * Schmid, M. F. & Booth, C. R. Methods for aligning and for averaging 3D volumes with missing data. _J. Struct. Biol._ 161, 243–248 (2008) Article
Google Scholar * Raytcheva, D. A., Haase-Pettingell, C., Piret, J. M. & King, J. A. Intracellular assembly of cyanophage Syn5 proceeds through a scaffold-containing procapsid. _J.
Virol._ 85, 2406–2415 (2011) Article CAS Google Scholar * Chen, D. H. et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. _Proc. Natl Acad. Sci.
USA_ 108, 1355–1360 (2011) Article ADS CAS Google Scholar * Prevelige, P. E., Thomas, D. & King, J. Scaffolding protein regulates the polymerization of P22 coat subunits into
icosahedral shells _in vitro_ . _J. Mol. Biol._ 202, 743–757 (1988) Article CAS Google Scholar * Hegde, S., Padilla-Sanchez, V., Draper, B. & Rao, V. B. Portal-large terminase
interactions of the bacteriophage T4 DNA packaging machine implicate a molecular lever mechanism for coupling ATPase to DNA translocation. _J. Virol._ 86, 4046–4057 (2012) Article CAS
Google Scholar * Aksyuk, A. A. & Rossmann, M. G. Bacteriophage assembly. _Viruses_ 3, 172–203 (2011) Article CAS Google Scholar * Hendrix, R. W. Bacteriophage assembly. _Nature_ 277,
172–173 (1979) Article ADS CAS Google Scholar * Fuller, D. N. et al. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor
processivity, and capsid transformations. _J. Mol. Biol._ 373, 1113–1122 (2007) Article CAS Google Scholar * Machado, I. M. & Atsumi, S. Cyanobacterial biofuel production. _J.
Biotechnol._ 162, 50–56 (2012) Article CAS Google Scholar * Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visulasualization of three-dimensional image data using IMOD.
_J. Struct. Biol._ 116, 71–76 (1996) Article CAS Google Scholar * Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle
reconstructions. _J. Struct. Biol._ 128, 82–97 (1999) Article CAS Google Scholar * Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. _J. Struct. Biol._
157, 38–46 (2007) Article CAS Google Scholar * Schmid, M. F. et al. A tail-like assembly at the portal vertex in intact herpes simplex type-1 virions. _PLoS Pathog._ 8, e1002961 (2012)
Article CAS Google Scholar * Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron
cryomicroscopy. _J. Mol. Biol._ 333, 721–745 (2003) Article CAS Google Scholar * Henderson, R. et al. Outcome of the first electron microscopy validation task force meeting. _Structure_
20, 205–214 (2012) Article CAS Google Scholar * Baker, M. L. et al. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. _Proc. Natl
Acad. Sci. USA_ 110, 12301–12306 (2013) Article ADS CAS Google Scholar * Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. _J. Comput.
Chem._ 25, 1605–1612 (2004) Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This research was supported by grants from the Robert Welch Foundation (Q1242) and National
Institutes of Health (P41GM123832 to W.C.; AI0175208 and PN2EY016525 to W.C. and J.A.K.; GM080139 to S.J.L.; T15LM007093 through the Gulf Coast Consortia to W.D. and R.H.R.; T32GM007330
through the MSTP to R.H.R.). We thank J. G. Galaz-Montoya and R. N. Irobalieva for editing of the manuscript. AUTHOR INFORMATION Author notes * John Flanagan Present address: Present
address: FEI, 5350 Dawson Creek Drive, Hillsboro, Oregon 97124, USA., AUTHORS AND AFFILIATIONS * Verna and Marrs Mclean Department of Biochemistry and Molecular Biology, National Center for
Macromolecular Imaging, Baylor College of Medicine, Houston, 77030, Texas, USA Wei Dai, Caroline Fu, John Flanagan, Htet A. Khant, Xiangan Liu, Ryan H. Rochat, Steve J. Ludtke, Michael F.
Schmid & Wah Chiu * Department of Biology, Massachusetts Institute of Technology, Cambridge, 02139, Massachusetts, USA Desislava Raytcheva, Cameron Haase-Pettingell & Jonathan A.
King * Department of Biology, Northeastern University, Boston, 02115, Massachusetts, USA Desislava Raytcheva & Jacqueline Piret * Program in Structural and Computational Biology and
Molecular Biophysics, Baylor College of Medicine, Houston, 77030, Texas, USA Ryan H. Rochat, Steve J. Ludtke, Michael F. Schmid & Wah Chiu * National Institute for Physiological
Sciences, National Institutes of Natural Sciences, 5-1 Higashiyama, Myodaiji, Okazaki 444-8787, Japan , Kuniaki Nagayama Authors * Wei Dai View author publications You can also search for
this author inPubMed Google Scholar * Caroline Fu View author publications You can also search for this author inPubMed Google Scholar * Desislava Raytcheva View author publications You can
also search for this author inPubMed Google Scholar * John Flanagan View author publications You can also search for this author inPubMed Google Scholar * Htet A. Khant View author
publications You can also search for this author inPubMed Google Scholar * Xiangan Liu View author publications You can also search for this author inPubMed Google Scholar * Ryan H. Rochat
View author publications You can also search for this author inPubMed Google Scholar * Cameron Haase-Pettingell View author publications You can also search for this author inPubMed Google
Scholar * Jacqueline Piret View author publications You can also search for this author inPubMed Google Scholar * Steve J. Ludtke View author publications You can also search for this author
inPubMed Google Scholar * Kuniaki Nagayama View author publications You can also search for this author inPubMed Google Scholar * Michael F. Schmid View author publications You can also
search for this author inPubMed Google Scholar * Jonathan A. King View author publications You can also search for this author inPubMed Google Scholar * Wah Chiu View author publications You
can also search for this author inPubMed Google Scholar CONTRIBUTIONS W.D., D.R. and C.F. prepared the samples and conducted the infection experiments under the advice of C.H.-P. and J.P.
W.D. collected the image data and reconstructed the tomograms; C.F. and H.A.K. established the Zernike phase plate imaging conditions in the microscope; K.N. provided the phase plates for
imaging; R.H.R. performed the statistical analysis. W.D. and M.F.S. developed the imaging processing methods and solved the structures of the phage assembly intermediates; W.D. and X.L.
refined the structures; J.F. and S.J.L. developed the symmetry-search algorithm for subvolume alignment; W.D., M.F.S., J.A.K. and W.C. interpreted the structures and wrote the manuscript.
CORRESPONDING AUTHOR Correspondence to Wah Chiu. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA
FIGURE 1 ZPC IMPROVES CONTRAST OF CRYOET IMAGES AND REVEALS DETAILED STRUCTURAL FEATURES OF SYN5-INFECTED CELLS. A, A conventional EM image of a Syn5-infected WH8109 cell. B, A ZPC image of
the same cell as shown in A under the same imaging conditions. EXTENDED DATA FIGURE 2 ZPC-CRYOEM SINGLE-PARTICLE IMAGES OF BIOCHEMICALLY PURIFIED MATURE SYN5 PHAGE. The particles are shown
with the tail pointing down and the wavy horn pointing up. The tail fibres appear to have variable conformations. EXTENDED DATA FIGURE 3 GENERAL LINEAR MODELLING OF CELLULAR CARBOXYSOME
NUMBER WITH PROGRESSION OF INFECTION. The number of carboxysomes remains roughly constant as infection progresses, indicating that their variation does not correlate with progression of
infection. The solid line indicates linear regression; the dashed lines indicate the 95% confidence limits. SUPPLEMENTARY INFORMATION ZERNIKE PHASE CONTRAST TILT SERIES IMAGES OF A
SYN5-INFECTED WH8109 CELL AT AN INTERMEDIATE STAGE OF INFECTION Tilt series of a frozen, hydrated Syn5-infected WH8109 cell was collected manually with an electron energy of 200 kV under low
dose conditions on a 4kx4k CCD camera at 25,000× microscope magnification. The tilt angles ranged from -60° to 60° at 3° step increments. The accumulated electron exposure for the specimen
in this tilt series was 40-50 electrons/Å2. The sampling of the data was calibrated to be 4.52Å/pixel. (MOV 11476 kb) VOLUME RENDERING AND ANNOTATION OF THE TOMOGRAM SHOWN IN VIDEO S1 WITH
COLOUR REPRESENTATIONS AS IN FIG. 2C The tomogram was reconstructed using IMOD software. It is displayed both section-by-section and by volume rendering. Features are annotated by colours to
designate molecular components attached and inside the cells, including cell envelope, thylakoid membrane, carboxysome, P-granules, ribosomes, infecting phages and phage assembly
intermediates. (MOV 27885 kb) ZERNIKE PHASE CONTRAST TILT SERIES IMAGES OF A SYN5-INFECTED WH8109 CELL AT A LATE STAGE OF INFECTION WITH A RUPTURED CELL MEMBRANE Tilt series of frozen,
hydrated Syn5-infected WH8109 cell was collected manually with an electron energy of 200 kV under low dose conditions on a 4kx4k CCD camera at 25,000× microscope magnification. The tilt
angles ranged from -60° to 60° at 3° step increments. The accumulated electron exposure for the specimen in this tilt series was 40-50 electrons/Å2. The sampling of the data was calibrated
to be 4.52Å/pixel. (MOV 13083 kb) VOLUME RENDERING AND ANNOTATION OF THE TOMOGRAM SHOWN IN VIDEO S3 WITH COLOUR REPRESENTATIONS AS IN FIG. 1 The tomogram was reconstructed using IMOD
software. It is displayed both section-by-section and by volume rendering. Features are annotated by colours to designate molecular components attached and inside the cells, including cell
envelope, thylakoid membrane, carboxysome, P-granules, ribosomes, infecting phages and phage assembly intermediates. An increased number of DNA containing phage particles are visible, and
overall contrast is enhanced over that in video S2, because the cell is beginning to rupture at this late stage of infection. (MOV 28142 kb) POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1
POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 POWERPOINT SLIDE FOR FIG. 4 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Dai, W., Fu, C.,
Raytcheva, D. _et al._ Visualizing virus assembly intermediates inside marine cyanobacteria. _Nature_ 502, 707–710 (2013). https://doi.org/10.1038/nature12604 Download citation * Received:
31 May 2013 * Accepted: 27 August 2013 * Published: 09 October 2013 * Issue Date: 31 October 2013 * DOI: https://doi.org/10.1038/nature12604 SHARE THIS ARTICLE Anyone you share the following
link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature
SharedIt content-sharing initiative
