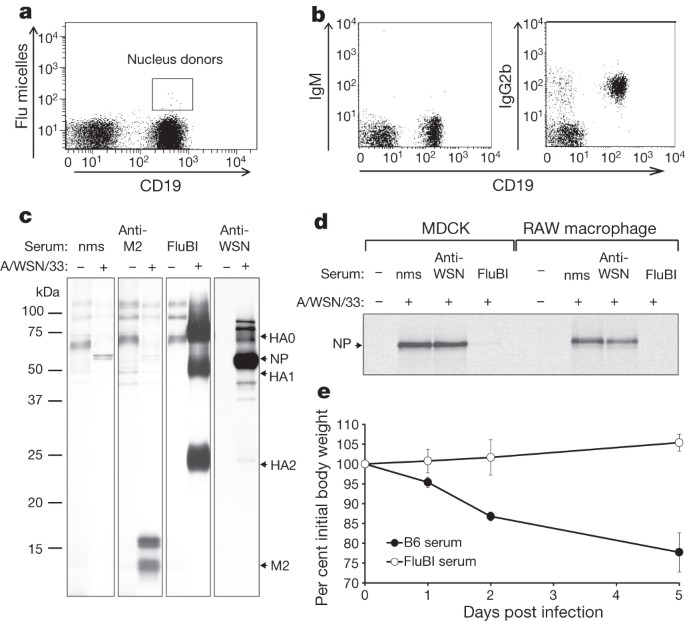
Antigen-specific b-cell receptor sensitizes b cells to infection by influenza virus
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Influenza A virus-specific B lymphocytes and the antibodies they produce protect against infection1. However, the outcome of interactions between an influenza
haemagglutinin-specific B cell via its receptor (BCR) and virus is unclear. Through somatic cell nuclear transfer we generated mice that harbour B cells with a BCR specific for the
haemagglutinin of influenza A/WSN/33 virus (FluBI mice). Their B cells secrete an immunoglobulin gamma 2b that neutralizes infectious virus. Whereas B cells from FluBI and control mice bind
equivalent amounts of virus through interaction of haemagglutinin with surface-disposed sialic acids, the A/WSN/33 virus infects only the haemagglutinin-specific B cells. Mere binding of
virus is not sufficient for infection of B cells: this requires interactions of the BCR with haemagglutinin, causing both disruption of antibody secretion and FluBI B-cell death within 18 h.
In mice infected with A/WSN/33, lung-resident FluBI B cells are infected by the virus, thus delaying the onset of protective antibody release into the lungs, whereas FluBI cells in the
draining lymph node are not infected and proliferate. We propose that influenza targets and kills influenza-specific B cells in the lung, thus allowing the virus to gain purchase before the
initiation of an effective adaptive response. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access
through your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS ACTIVATION AND MATURATION OF ANTIGEN-SPECIFIC B CELLS IN NONECTOPIC LUNG INFILTRATES ARE
INDEPENDENT OF GERMINAL CENTER REACTIONS IN THE DRAINING LYMPH NODE Article Open access 11 April 2025 HUMAN NAÏVE B CELLS SHOW EVIDENCE OF ANERGY AND CLONAL REDEMPTION FOLLOWING VACCINATION
Article Open access 14 May 2025 INFLUENZA VIRUS INFECTION EXPANDS THE BREADTH OF ANTIBODY RESPONSES THROUGH IL-4 SIGNALLING IN B CELLS Article Open access 18 June 2021 ACCESSION CODES
PRIMARY ACCESSIONS GENBANK/EMBL/DDBJ * KF419287 * KF419288) REFERENCED ACCESSIONS GENBANK/EMBL/DDBJ * KF419287 * KF419288 DATA DEPOSITS Sequences of the mouse _V_, _D_ and _J_ genes were
deposited in GenBank with accession numbers KF419287 and KF419288. REFERENCES * Valkenburg, S. A. et al. Immunity to seasonal and pandemic influenza A viruses. _Microbes Infect._ 13, 489–501
(2011) Article CAS Google Scholar * Onodera, T. et al. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. _Proc. Natl
Acad. Sci. USA_ 109, 2485–2490 (2012) Article ADS CAS Google Scholar * Manicassamy, B. et al. Analysis of _in vivo_ dynamics of influenza virus infection in mice using a GFP reporter
virus. _Proc. Natl Acad. Sci. USA_ 107, 11531–11536 (2010) Article ADS CAS Google Scholar * Popp, M. W., Karssemeijer, R. A. & Ploegh, H. L. Chemoenzymatic site-specific labeling of
influenza glycoproteins as a tool to observe virus budding in real time. _PLoS Pathog._ 8, e1002604 (2012) Article CAS Google Scholar * Popp, M. W. & Ploegh, H. L. Making and breaking
peptide bonds: protein engineering using sortase. _Angew. Chem. Int. Edn Engl._ 50, 5024–5032 (2011) Article CAS Google Scholar * Antos, J. M., Miller, G. M., Grotenbreg, G. M. &
Ploegh, H. L. Lipid modification of proteins through sortase-catalyzed transpeptidation. _J. Am. Chem. Soc._ 130, 16338–16343 (2008) Article CAS Google Scholar * Dougan, S. K. et al.
IgG1+ ovalbumin-specific B-cell transnuclear mice show class switch recombination in rare allelically included B cells. _Proc. Natl Acad. Sci. USA_ 109, 13739–13744 (2012) Article ADS CAS
Google Scholar * Kirak, O. et al. Transnuclear mice with predefined T cell receptor specificities against _Toxoplasma gondii_ obtained via SCNT. _Science_ 328, 243–248 (2010) Article ADS
CAS Google Scholar * Dougan, S. K. et al. Transnuclear TRP1-specific CD8 T cells with high or low affinity TCRs show equivalent anti-tumor activity. _Cancer Immunol. Res._ 1, 99–111
(2013) Article CAS Google Scholar * Wiley, D. C. & Skehel, J. J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. _Annu. Rev. Biochem._ 56,
365–394 (1987) Article CAS Google Scholar * Stray, S. J., Cummings, R. D. & Air, G. M. Influenza virus infection of desialylated cells. _Glycobiology_ 10, 649–658 (2000) Article CAS
Google Scholar * Thompson, C. I., Barclay, W. S., Zambon, M. C. & Pickles, R. J. Infection of human airway epithelium by human and avian strains of influenza a virus. _J. Virol._ 80,
8060–8068 (2006) Article CAS Google Scholar * Pedroso de Lima, M. C. et al. Target cell membrane sialic acid modulates both binding and fusion activity of influenza virus. _Biochim.
Biophys. Acta_ 1236, 323–330 (1995) Article Google Scholar * Huang, R. T., Lichtenberg, B. & Rick, O. Involvement of annexin V in the entry of influenza viruses and role of
phospholipids in infection. _FEBS Lett._ 392, 59–62 (1996) Article CAS Google Scholar * Chu, V. C. & Whittaker, G. R. Influenza virus entry and infection require host cell N-linked
glycoprotein. _Proc. Natl Acad. Sci. USA_ 101, 18153–18158 (2004) Article ADS CAS Google Scholar * Londrigan, S. L. et al. N-linked glycosylation facilitates sialic acid-independent
attachment and entry of influenza A viruses into cells expressing DC-SIGN or L-SIGN. _J. Virol._ 85, 2990–3000 (2011) Article CAS Google Scholar * Eierhoff, T., Hrincius, E. R., Rescher,
U., Ludwig, S. & Ehrhardt, C. The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. _PLoS Pathog._ 6, e1001099 (2010) Article Google
Scholar * Harwood, N. E. & Batista, F. D. Early events in B cell activation. _Annu. Rev. Immunol._ 28, 185–210 (2010) Article CAS Google Scholar * Witte, M. D. et al. Preparation of
unnatural N-to-N and C-to-C protein fusions. _Proc. Natl Acad. Sci. USA_ 109, 11993–11998 (2012) Article ADS CAS Google Scholar * Moltedo, B. et al. Cutting edge: stealth influenza virus
replication precedes the initiation of adaptive immunity. _J. Immunol._ 183, 3569–3573 (2009) Article CAS Google Scholar * Moltedo, B., Li, W., Yount, J. S. & Moran, T. M. Unique
type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. _PLoS Pathog._ 7, e1002345 (2011) Article CAS Google Scholar
* Joo, H. M., He, Y. & Sangster, M. Y. Broad dispersion and lung localization of virus-specific memory B cells induced by influenza pneumonia. _Proc. Natl Acad. Sci. USA_ 105, 3485–3490
(2008) Article ADS CAS Google Scholar * Jones, P. D. & Ada, G. L. Influenza-specific antibody-secreting cells and B cell memory in the murine lung after immunization with wild-type,
cold-adapted variant and inactivated influenza viruses. _Vaccine_ 5, 244–248 (1987) Article CAS Google Scholar * Popp, M. W., Antos, J. M. & Ploegh, H. L. Site-specific protein
labeling via sortase-mediated transpeptidation. _Curr. Protoc. Protein Sci._ 15, Unit 15.13. (2009) * Boes, M. et al. T-cell engagement of dendritic cells rapidly rearranges MHC class II
transport. _Nature_ 418, 983–988 (2002) Article ADS CAS Google Scholar * Morgan, D. J., McLain, L. & Dimmock, N. J. Protection of three strains of mice against lethal influenza in
vivo by defective interfering virus. _Virus Res._ 29, 179–193 (1993) Article CAS Google Scholar * Kirak, O. et al. Transnuclear mice with pre-defined T cell receptor specificities against
_Toxoplasma gondii_ obtained via SCNT. _J. Vis. Exp._ 43, e2168 (2010) Google Scholar * Sehrawat, S. et al. CD8+ T cells from mice transnuclear for a TCR that recognizes a single
H-2Kb-restricted MHV68 epitope derived from gB-ORF8 help control infection. _Cell Rep._ 1, 461–471 (2012) Article CAS Google Scholar * Kishigami, S. et al. Production of cloned mice by
somatic cell nuclear transfer. _Nature Protocols_ 1, 125–138 (2006) Article CAS Google Scholar * Simons, K., Helenius, A., Leonard, K., Sarvas, M. & Gething, M. J. Formation of
protein micelles from amphiphilic membrane proteins. _Proc. Natl Acad. Sci. USA_ 75, 5306–5310 (1978) Article ADS CAS Google Scholar Download references ACKNOWLEDGEMENTS S.K.D. and C.G.
were funded by the Cancer Research Institute. J.J.C. was funded by the Human Frontiers Science Program. F.W.A., R.J. and H.L.P. are funded by grants from the National Institutes of Health.
F.W.A. is a Howard Hughes Medical Institute investigator. S.K.D. and H.L.P. are funded by the American Association for Cancer Research-Pancreatic Cancer Action Network. We are grateful to P.
Wisniewski for cell sorting, to J. Jackson for mouse husbandry, to G. Bell for statistical analysis, to N. Watson for electron microscopy and to M. Witte for sortase nucleophiles. AUTHOR
INFORMATION Author notes * Stephanie K. Dougan and Joseph Ashour: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Whitehead Institute for Biomedical Research, 9
Cambridge Center, Cambridge, 02142, Massachusetts, USA Stephanie K. Dougan, Joseph Ashour, Roos A. Karssemeijer, Maximilian W. Popp, Ana M. Avalos, Marta Barisa, Arwen F. Altenburg, Jessica
R. Ingram, Juan Jose Cragnolini, Rudolf Jaenisch & Hidde L. Ploegh * Department of Biology, Massachusetts Institute of Technology, Cambridge, 02139, Massachusetts, USA Maximilian W. Popp
& Hidde L. Ploegh * Boston Children’s Hospital, Karp Family Research Building, One Blackfan Circle Boston, 02115, Massachusetts, USA Chunguang Guo & Frederick W. Alt Authors *
Stephanie K. Dougan View author publications You can also search for this author inPubMed Google Scholar * Joseph Ashour View author publications You can also search for this author inPubMed
Google Scholar * Roos A. Karssemeijer View author publications You can also search for this author inPubMed Google Scholar * Maximilian W. Popp View author publications You can also search
for this author inPubMed Google Scholar * Ana M. Avalos View author publications You can also search for this author inPubMed Google Scholar * Marta Barisa View author publications You can
also search for this author inPubMed Google Scholar * Arwen F. Altenburg View author publications You can also search for this author inPubMed Google Scholar * Jessica R. Ingram View author
publications You can also search for this author inPubMed Google Scholar * Juan Jose Cragnolini View author publications You can also search for this author inPubMed Google Scholar *
Chunguang Guo View author publications You can also search for this author inPubMed Google Scholar * Frederick W. Alt View author publications You can also search for this author inPubMed
Google Scholar * Rudolf Jaenisch View author publications You can also search for this author inPubMed Google Scholar * Hidde L. Ploegh View author publications You can also search for this
author inPubMed Google Scholar CONTRIBUTIONS S.K.D. and J.A. contributed equally. Somatic cell nuclear transfer was performed by S.K.D.; S.K.D., J.A., R.A.K., M.W.P., M.B. and A.F.A.
performed experiments. A.M.A. generated hybridomas. J.R.I. and J.J.C. generated flu-specific VHHs. C.G. sequenced the BCR loci. F.W.A. and R.J. provided advice and reagents. S.K.D., J.A. and
H.L.P. designed experiments, analysed data and wrote the paper. CORRESPONDING AUTHOR Correspondence to Hidde L. Ploegh. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing financial interests. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIGURE 1 FLU MICELLES STAIN HA-SPECIFIC B CELLS. A, Schematic for preparation of glycoprotein micelles from
HA–SRTAlexa 647 virus. B, Immunoprecipitation of HA–Alexa 647 with anti- Alexa 647 monoclonal antibody. Triton X100-disrupted virions were incubated with 400 μg anti-Alexa 647 overnight and
HA–Alexa 647 was then recovered using protein G-Sepharose. Bound proteins were eluted with 0.1 M glycine pH 2.8. W, wash; E, elution. C, Typhoon image of the fractions obtained from a linear
sucrose gradient after 20 h centrifugation (107,900_g_). D, Fraction 8 from the sucrose gradient was concentrated and sucrose-depleted by centrifugation over a 30 kDa filter (Amicon
UltraCel). The preparation was stained with phosphotungstate and examined by transmission electron microscopy (×150,000 magnification). E, Splenocytes from mice infected with A/WSN/33 or
control mice were stained with anti-CD19 and HA–Alexa 647 micelles and analysed by cytofluorometry. Plots are representative of 6 mice per group. EXTENDED DATA FIGURE 2 FLUBI ANTIBODY IS OF
THE IGG2B SUBCLASS. ELISA plates were coated with A/WSN/33-infected MDCK cell lysate and exposed to 1:100 diluted serum from a single C57BL/6 (wt), FluBI, FluBI;_Rag2_−/−, or wild-type mouse
infected with A/WSN/33. Plates were washed and probed with isotype-specific secondary antibodies. Uninfected wild-type mice have flu-reactive antibodies of the IgM subclass. Flu-specific
IgE was not detected in any sample. Error bars are s.d. of samples analysed in triplicate. EXTENDED DATA FIGURE 3 SEQUENCE OF THE VDJ AND VJ SEGMENTS OF THE FLUBI ANTIBODY. Genomic DNA was
prepared from tails of FluBI mice. The heavy and light chain rearrangements were first identified by amplifying and sequencing of the segments with degenerate primers: for heavy chain:
forward 5′-ARGCCTGGGRCTTCAGTGAAG-3′ and reverse 5′-AGGCTCTGAGATCCCTAGACAG-3′; for light chain: forward 5′-GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC-3′ and reverse
5′-ATGCGACGTCAACTGATAATGAGCCCTCTCC-3′. Then the full sequences of the rearranged heavy and light chain segments were obtained using specific primers: forward 5′-TTACTGAGCACACAGGACCTC-3′ and
reverse 5′-AGGCTCTGAGATCCCTAGACAG-3′; for light chain: forward 5′-CAGCCCATATTCTCCCATGT-3′ and reverse 5′-ATGCGACGTCAACTGATAATGAGCCCTCTCC-3′. Amplified products were agarose gel-purified and
sequenced. Sequences were aligned to the NCBI mouse _V_, _D_ and _J_ genes using IgBlast. Sequences were deposited in GenBank (accession numbers KF419287 and KF419288). EXTENDED DATA FIGURE
4 FLUBI MICE LACK B-1A B CELLS, BUT SHOW NEAR-NORMAL DEVELOPMENT OF FOLLICULAR B CELLS. Cells were isolated from spleen, lymph node (LN, pooled mesenteric and cervical), peritoneal cavity
and bone marrow of FluBI, FluBI _Rag2_−/− or C57BL/6 mice. Erythrocytes were lysed and cells were stained with the indicated antibodies and 7-AAD viability dye. LN plots were gated on total
live cells. All other populations were gated on CD19+ live cells. Numbers indicated the percentage of cells in the indicated gates. B-1a B cells (CD5+) are absent and B-1b B cells
(CD5−CD11b+) are reduced in the peritoneal cavity of FluBI and FluBI _Rag2_−/− mice. Plots are representative of 5 mice per group. EXTENDED DATA FIGURE 5 FLUBI B CELLS ARE INFECTED BY
A/WSN/33. CD40-activated OBI or FluBI B cells were incubated with A/WSN/33 virus at an MOI of 1.0 for 30 min on ice. Cells were then washed and incubated at 37 °C in RPMI (0.2% BSA). At 2
h.p.i., cells were fixed, permeabilized and stained with anti-IgG and TAMRA-conjugated anti-NP (VHH54, derived from alpaca; see Extended Data Fig. 9). A, Cells were visualized by confocal
microscopy. B, Cells from A were scored as VHH54-positive or -negative. Error bars represent s.d. of positive cells counted per field (3 fields counted; ∼200 total cells were counted per
group). EXTENDED DATA FIGURE 6 ANTIBODY SECRETED BY FLUBI B CELLS DOES NOT CROSS-REACT WITH OTHER STRAINS OF INFLUENZA VIRUS. ELISA plates were coated with A/WSN/33 (H1N1), A/Udorn/307/1972
(H3N2) or A/Puerto Rico/8/1934 (H1N1) overnight at 4°. Plates were then washed, blocked with 10% fetal bovine serum and exposed to FluBI hybridoma supernatant or WSN-infected serum at the
indicated dilutions. Bound antibody was detected using horseradish peroxidase-coupled anti-IgG2b secondary reagent. EXTENDED DATA FIGURE 7 FLUBI B CELLS ARE NOT INFECTED WITH A/PUERTO
RICO/8/1934 VIRUS _IN VIVO_. C57BL/6 mice were administered 5 × 106 MHCII–GFP+ FluBI B cells 2 h before intranasal infection with 2 × 105 p.f.u. per mouse of either A/WSN/33 (WSN) or
A/Puerto Rico/8/1934 (PR8). Mice were euthanized 3 days post-infection, and lung resident cells were stained with anti-CD19 and TAMRA-conjugated VHH68 (anti-HA) or TAMRA-conjugated VHH52/54
(anti-NP). A, Representative plots gated on CD19+ cells. B, Quantification of flu-antigen positive cells as shown in A. _n_ = 3. Error bars are s.d. p = 0.06 using two-sided _t_-test.
EXTENDED DATA FIGURE 8 PROLIFERATING FLUBI CELLS IN THE MEDIASTINAL LYMPH NODE ARE PLASMABLASTS. A, Mediastinal lymph node cells from day 6 post live infection mice described in Fig. 4 were
analysed by confocal microscopy. GFP+ cells displayed a morphology consistent with plasmablasts. B, MSLN cells from day 6 post live infection mice described in Figure 4 were analysed by
cytofluorometry. Proliferating (violet low) cells were B220low and CD138+. EXTENDED DATA FIGURE 9 ALPACA-DERIVED VHHS RECOGNIZE HA AND NP FROM A/WSN/33. A, An alpaca was immunized with
ethanol-fixed influenza virus. Phage display libraries were constructed from selectively amplified VHH-specific complementary DNA using peripheral blood lymphocytes as starting material, and
panned twice against sortase labelled influenza HA–SRTbiotin virus bound to streptavidin coupled beads. VHH sequences obtained from specific binders were expressed with a sortase
recognition motif to allow direct conjugation of biotin or fluorophores. B, VHH54 and VHH68 conjugated directly to agarose beads were used to precipitate lysates of A/WSN/33 infected,
[35S]cysteine/methionine-labelled MDCK cells. EXTENDED DATA FIGURE 10 FLU-SPECIFIC VHHS CAN STAIN INFECTED FLUBI B CELLS. B cells from OBI or FluBI mice were cultured for 24 h in RPMI
containing anti-CD40 (1 μg ml−1) before exposure to A/WSN/33. OBI B cells, FluBI B cells and MDCK cells were incubated with A/WSN/33 at an MOI of 1.0 for 30 min on ice, washed once with PBS,
and transferred to 37 °C in RPMI (0.2% BSA). At 5 h post infection, cells were washed, permeabilized, fixed and stained using TAMRA-conjugated flu-specific VHHs (1 μg in 50 μl). Infected
MDCK cells were analysed in parallel as a positive control. Cells were analysed by cytofluorometry using a BD Fortessa. POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1 POWERPOINT SLIDE FOR
FIG. 2 POWERPOINT SLIDE FOR FIG. 3 POWERPOINT SLIDE FOR FIG. 4 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Dougan, S., Ashour, J., Karssemeijer, R.
_et al._ Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. _Nature_ 503, 406–409 (2013). https://doi.org/10.1038/nature12637 Download citation * Received:
24 May 2013 * Accepted: 04 September 2013 * Published: 20 October 2013 * Issue Date: 21 November 2013 * DOI: https://doi.org/10.1038/nature12637 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative
