Single-cell topological RNA-seq analysis reveals insights into cellular differentiation and development
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

Transcriptional programs control cellular lineage commitment and differentiation during development. Understanding of cell fate has been advanced by studying single-cell RNA-sequencing
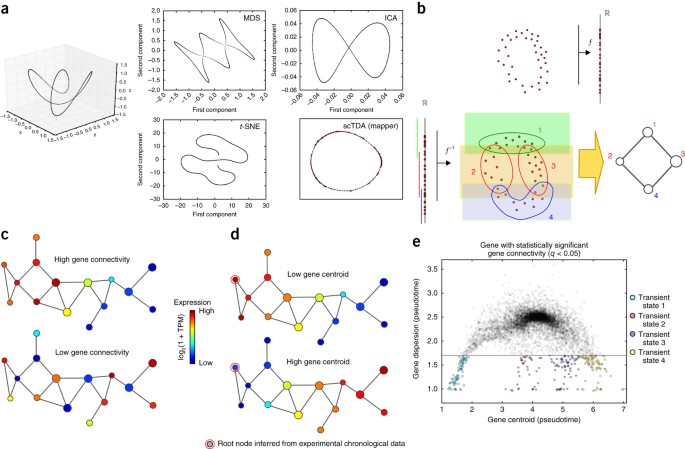
(RNA-seq) but is limited by the assumptions of current analytic methods regarding the structure of data. We present single-cell topological data analysis (scTDA), an algorithm for
topology-based computational analyses to study temporal, unbiased transcriptional regulation. Unlike other methods, scTDA is a nonlinear, model-independent, unsupervised statistical
framework that can characterize transient cellular states. We applied scTDA to the analysis of murine embryonic stem cell (mESC) differentiation in vitro in response to inducers of motor
neuron differentiation. scTDA resolved asynchrony and continuity in cellular identity over time and identified four transient states (pluripotent, precursor, progenitor, and fully
differentiated cells) based on changes in stage-dependent combinations of transcription factors, RNA-binding proteins, and long noncoding RNAs (lncRNAs). scTDA can be applied to study
asynchronous cellular responses to either developmental cues or environmental perturbations.
We thank T. Jessell, N. Francis, and H. Phatnani for critical reading of the manuscript. A.H.R. and T.M. thank the New York Genome Center and D. Goldstein for sequencing support, S. Morton
for providing Engrailed antibody, and P. Sims for experimental discussions. P.G.C. and R.R. thank A.J. Levine, G. Carlsson, F. Abate, I. Filip, S. Zairis, U. Rubin, and P. van Nieuwenhuizen
for useful comments and discussions, O.T. Elliott for technical support with the online database, and Ayasdi Inc. for technical support. The work of P.G.C. and R.R. is supported by the NIH
grants U54-CA193313-01 and R01GM117591. The work of A.H.R., E.K.K., T.J.R. and T.M. is supported by ALS Therapy Alliance grant ATA-2013-F-056 and NIH grant NS088992.
Abbas H Rizvi and Pablo G Camara: These authors contributed equally to this work.
Department of Biochemistry and Molecular Biophysics, Columbia University Medical Center, New York, New York, USA
Abbas H Rizvi, Elena K Kandror, Thomas J Roberts & Tom Maniatis
The Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, New York, USA
Abbas H Rizvi, Elena K Kandror, Thomas J Roberts, Ira Schieren & Tom Maniatis
Department of Systems Biology, Columbia University Medical Center, New York, New York, USA
Department of Biomedical Informatics, Columbia University Medical Center, New York, New York, USA
Howard Hughes Medical Institute, Columbia University Medical Center, New York, New York, USA
P.G.C. and R.R. developed the topology-based computational approach (scTDA) and applied it to single cell RNA sequencing data. A.H.R., E.K.K., and T.M. designed all experiments. A.H.R.,
E.K.K., and T.J.R. conducted experiments. I.S. conducted all flow cytometry. A.H.R., P.G.C., E.K.K., T.M., and R.R. analyzed the data and wrote the manuscript.
Supplementary Figures 1–20, Supplementary Table 1, and Supplementary Notes 1 and 2 (PDF 9143 kb)
All genes characterization of the expression profile in the topological representation of 80 embryonic (E18.5) mouse lung 35 epithelial cells. (XLSX 542 kb)
Characterization of the expression profile in the topological representation of 1,529 individual cells from 88 human preimplantation embryos. (XLSX 1929 kb)
All genes characterization of the expression profile in the topological representation of 272 newborn neurons from the mouse neocortex. (XLSX 1660 kb)
Barcoded reverse transcription primers utilized in motor neuron differentiation experiment 2. (XLSX 5 kb)
Python code for single-cell topological data analysis (scTDA). Also available at https://github.com/RabadanLab/scTDA. (TXT 94 kb)
Anyone you share the following link with will be able to read this content:
