Multiplexed detection of pathogen dna with dna-based fluorescence nanobarcodes
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Rapid, multiplexed, sensitive and specific molecular detection is of great demand in gene profiling, drug screening, clinical diagnostics and environmental analysis1,2,3. One of the
major challenges in multiplexed analysis is to identify each specific reaction with a distinct label or 'code'4. Two encoding strategies are currently used: positional encoding,
in which every potential reaction is preassigned a particular position on a solid-phase support such as a DNA microarray5,6,7,8, and reaction encoding, where every possible reaction is
uniquely tagged with a code that is most often optical or particle based4,9,10,11,12,13. The micrometer size, polydispersity, complex fabrication process and nonbiocompatibility of current
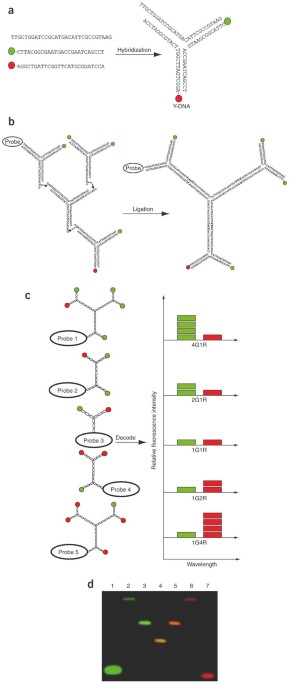
codes limit their usability1,4,12. Here we demonstrate the synthesis of dendrimer-like DNA-based, fluorescence-intensity-coded nanobarcodes, which contain a built-in code and a probe for
molecular recognition. Their application to multiplexed detection of the DNA of several pathogens is first shown using fluorescence microscopy and dot blotting, and further demonstrated
using flow cytometry that resulted in detection that was sensitive (attomole) and rapid. Access through your institution Buy or subscribe This is a preview of subscription content, access
via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy
this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: *
Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS A UNIVERSAL REAGENT FOR DETECTION OF EMERGING DISEASES
USING BIOENGINEERED MULTIFUNCTIONAL YEAST NANOFRAGMENTS Article 08 June 2023 ULTRA-SENSITIVE AND RAPID DETECTION OF NUCLEIC ACIDS AND MICROORGANISMS IN BODY FLUIDS USING SINGLE-MOLECULE
TETHERING Article Open access 22 September 2020 PROGRAMMING BACTERIA FOR MULTIPLEXED DNA DETECTION Article Open access 10 April 2023 REFERENCES * Han, M., Gao, X.H., Su, J.Z. & Nie, S.
Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. _Nat. Biotechnol._ 19, 631–635 (2001). Article CAS Google Scholar * Fulton, R.J., McDade, R.L., Smith, P.L.,
Kienker, L.J. & Kettman, J.R. Advanced multiplexed analysis with the FlowMetrix(TM) system. _Clin. Chem._ 43, 1749–1756 (1997). Article CAS Google Scholar * Steemers, F.J., Ferguson,
J.A. & Walt, D.R. Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. _Nat. Biotechnol._ 18, 91–94 (2000). Article CAS Google Scholar * Braeckmans, K., De
Smedt, S.C., Leblans, M., Pauwels, R. & Demeester, J. Encoding microcarriers: Present and future technologies. _Nat. Rev. Drug Discov._ 1, 447–456 (2002). Article CAS Google Scholar *
Duggan, D., Bittner, M., Chen, Y., Meltzer, P. & Trent, J. Expression profiling using cDNA microarrays. _Nat. Genet._ 21, 10–14 (1999). Article CAS Google Scholar * DeRisi, J. et al.
Use of a cDNA microarray to analyse gene expression patterns in human cancer. _Nat. Genet._ 14, 457–460 (1996). Article CAS Google Scholar * Schena, M., Shalon, D., Davis, R. &
Brown, P. Quantitative monitoring of gene-expression patterns with a complementary-DNA microarray. _Science_ 270, 467–470 (1995). Article CAS Google Scholar * Cheung, V.G. et al. Making
and reading microarrays. _Nat. Genet._ 21, 15–19 (1999). Article CAS Google Scholar * Cunin, F. et al. Biomolecular screening with encoded porous-silicon photonic crystals. _Nat. Mater._
1, 39–41 (2002). Article CAS Google Scholar * Wang, J., Liu, G.D. & Rivas, G. Encoded beads for electrochemical identification. _Anal. Chem._ 75, 4667–4671 (2003). Article CAS
Google Scholar * Chan, W.C. et al. Luminescent quantum dots for multiplexed biological detection and imaging. _Curr. Opin. Biotechnol._ 13, 40–46 (2002). Article CAS Google Scholar *
Nicewarner-Pena, S.R. et al. Submicrometer metallic barcodes. _Science_ 294, 137–141 (2001). Article CAS Google Scholar * Nam, J.M., Thaxton, C.S. & Mirkin, C.A. Nanoparticle-based
bio-bar codes for the ultrasensitive detection of proteins. _Science_ 301, 1884–1886 (2003). Article CAS Google Scholar * Li, Y. et al. Controlled assembly of dendrimer-like DNA. _Nat.
Mater._ 3, 38–42 (2004). Article CAS Google Scholar * Luo, D. The road from biology to materials. _Mater. Today_ 6, 38–43 (2003). Article CAS Google Scholar * Ried, T., Baldini, A.,
Rand, T.C. & Ward, D.C. Simultaneous visualization of 7 different DNA probes by _in situ_ hybridization using combinatorial fluorescence and digital imaging microscopy. _Proc. Natl.
Acad. Sci. USA_ 89, 1388–1392 (1992). Article CAS Google Scholar * Taton, T., Mirkin, C. & Letsinger, R. Scanometric DNA array detection with nanoparticle probes. _Science_ 289,
1757–1760 (2000). Article CAS Google Scholar * Sjostedt, A., Eriksson, U., Berglund, L. & Tarnvik, A. Detection of _Francisella tularensis_ in ulcers of patients with tularemia by
PCR. _J. Clin. Microbiol._ 35, 1045–1048 (1997). Article CAS Google Scholar * Sanchez, A. et al. Detection and molecular characterization of Ebola viruses causing disease in human and
nonhuman primates. _J. Infect. Dis._ 179, S164–S169 (1999). Article CAS Google Scholar * Poon, L.L. et al. Detection of SARS coronavirus in patients with severe acute respiratory syndrome
by conventional and real-time quantitative reverse transcription-PCR assays. _Clin. Chem._ 50, 67–72 (2004). Article CAS Google Scholar * Fuja, T., Hou, S. & Bryant, P. A multiplex
microsphere bead assay for comparative RNA expression analysis using flow cytometry. _J. Biotechnol._ 108, 193–205 (2004). Article CAS Google Scholar * Vignali, D.A. Multiplexed
particle-based flow cytometric assays. _J. Immunol. Methods_ 243, 243–255 (2000). Article CAS Google Scholar * Wang, J. et al. Adsorption and detection of DNA dendrimers at carbon
electrodes. _Electroanal._ 10, 553–556 (1998). Article CAS Google Scholar * Wang, J., Jiang, M., Nilsen, T. & Getts, R. Dendritic nucleic acid probes for DNA biosensors. _J. Am. Chem.
Soc._ 120, 8281–8282 (1998). Article CAS Google Scholar * Nilsen, T., Grayzel, J. & Prensky, W. Dendritic nucleic acid structures. _J. Theor. Biol._ 187, 273–284 (1997). Article CAS
Google Scholar * Capaldi, S., Getts, R.C. & Jayasena, S.D. Signal amplification through nucleotide extension and excision on a dendritic DNA platform. _Nucleic Acids Res._ 28, e21
(2000). Article CAS Google Scholar * Lowe, M., Spiro, A., Zhang, Y. & Getts, R. Multiplexed, particle-based detection of DNA using flow cytometry with 3DNA dendrimers for signal
amplification. _Cytom_ Part A 60A, 135–144 (2004). Article CAS Google Scholar * Shchepinov, M., Mir, K., Elder, J., Frank-Kamenetskii, M. & Southern, E. Oligonucleotide dendrimers:
stable nano-structures. _Nucleic Acids Res._ 27, 3035–3041 (1999). Article CAS Google Scholar * Shchepinov, M., Udalova, I., Bridgman, A. & Southern, E. Oligonucleotide dendrimers:
synthesis and use as polylabelled DNA probes. _Nucleic Acids Res._ 25, 4447–4454 (1997). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We wish to acknowledge the Cornell
Center for Advanced Technology and the Cornell Center for Vertebrate Genomics for financial support. This material is based upon work supported in part by the Science and Technology Center
Program of the National Science Foundation under agreement no. ECS-9876771. We thank Yung-Fu Chang for providing pathogen genomic DNA (_Mycobacterium avium_ subsp. _paratuberculosis_) and
Carol Bayles for technical support on microscopy. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Biological and Environmental Engineering, Cornell University, Ithaca,
14853-5701, New York, USA Yougen Li, Yen Thi Hong Cu & Dan Luo Authors * Yougen Li View author publications You can also search for this author inPubMed Google Scholar * Yen Thi Hong Cu
View author publications You can also search for this author inPubMed Google Scholar * Dan Luo View author publications You can also search for this author inPubMed Google Scholar
CORRESPONDING AUTHOR Correspondence to Dan Luo. ETHICS DECLARATIONS COMPETING INTERESTS A patent for similar technology is being filed, the value of which may be increased by this
publication. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIG. 1 Evaluation of nanobarcodes with agarose gel electrophoresis. (PDF 745 kb) SUPPLEMENTARY FIG. 2 Schematic drawing of nanobarcode
denaturation (without showing fluorescence dyes). (PDF 96 kb) SUPPLEMENTARY FIG. 3 DNA nanobarcode quantitative decoding based on microbead populations. (PDF 42 kb) SUPPLEMENTARY FIG. 4
Statistics multiplexed DNA detection using flow cytometry. (PDF 55 kb) SUPPLEMENTARY TABLE 1 Building oligonucleotides (PDF 47 kb) SUPPLEMENTARY TABLE 2 Capture probes, report probes and
target DNA (PDF 47 kb) SUPPLEMENTARY TABLE 3 Y-DNA building blocks (PDF 43 kb) SUPPLEMENTARY TABLE 4 DNA nanobarcodes (PDF 40 kb) SUPPLEMENTARY TABLE 5 Code library (PDF 45 kb) SUPPLEMENTARY
NOTE 1 (PDF 66 KB) SUPPLEMENTARY NOTE 2 (PDF 42 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Li, Y., Cu, Y. & Luo, D. Multiplexed detection
of pathogen DNA with DNA-based fluorescence nanobarcodes. _Nat Biotechnol_ 23, 885–889 (2005). https://doi.org/10.1038/nbt1106 Download citation * Received: 22 February 2005 * Accepted: 05
May 2005 * Published: 01 July 2005 * Issue Date: 01 July 2005 * DOI: https://doi.org/10.1038/nbt1106 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
