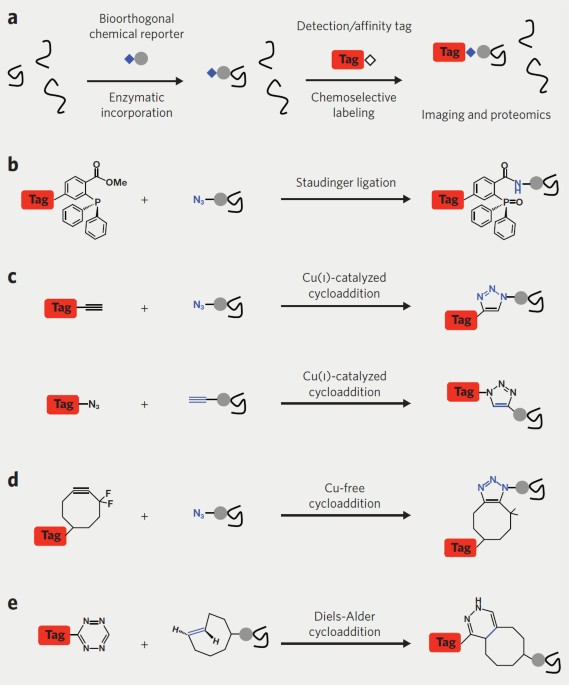
Chemical reporters for biological discovery
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Functional tools are needed to understand complex biological systems. Here we review how chemical reporters in conjunction with bioorthogonal labeling methods can be used to image
and retrieve nucleic acids, proteins, glycans, lipids and other metabolites _in vitro_, in cells as well as in whole organisms. By tagging these biomolecules, researchers can now monitor
their dynamics in living systems and discover specific substrates of cellular pathways. These advances in chemical biology are thus providing important tools to characterize biological
pathways and are poised to facilitate our understanding of human diseases. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this
article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in
* Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS DEVELOPING BIOORTHOGONAL PROBES TO SPAN A SPECTRUM OF REACTIVITIES
Article 21 July 2020 CHARTING UNKNOWN METABOLIC REACTIONS BY MASS SPECTROMETRY-RESOLVED STABLE-ISOTOPE TRACING METABOLOMICS Article Open access 31 May 2025 EXPANDING CHEMISTRY THROUGH IN
VITRO AND IN VIVO BIOCATALYSIS Article 03 July 2024 CHANGE HISTORY * _ 19 DECEMBER 2013 In the version of this Review Article initially published, a double bond was missing from the chemical
structure of the cyclooctene on the left side of the reaction arrow in Figure 1e. The chemical structure has been corrected in the HTML and PDF versions of the article. _ REFERENCES *
Prescher, J.A. & Bertozzi, C.R. Chemistry in living systems. _Nat. Chem. Biol._ 1, 13–21 (2005). Article CAS PubMed Google Scholar * Sletten, E.M. & Bertozzi, C.R. From mechanism
to mouse: a tale of two bioorthogonal reactions. _Acc. Chem. Res._ 44, 666–676 (2011). Article CAS PubMed PubMed Central Google Scholar * Nomura, D.K., Dix, M.M. & Cravatt, B.F.
Activity-based protein profiling for biochemical pathway discovery in cancer. _Nat. Rev. Cancer_ 10, 630–638 (2010). Article CAS PubMed PubMed Central Google Scholar * Salic, A. &
Mitchison, T.J. A chemical method for fast and sensitive detection of DNA synthesis _in vivo_. _Proc. Natl. Acad. Sci. USA_ 105, 2415–2420 (2008). THIS STUDY DESCRIBES THE FIRST ALKYNE
CHEMICAL REPORTER FOR LABELING NUCLEIC ACIDS IN CELLS AND ANIMALS. Article CAS PubMed PubMed Central Google Scholar * Jao, C.Y. & Salic, A. Exploring RNA transcription and turnover
_in vivo_ by using click chemistry. _Proc. Natl. Acad. Sci. USA_ 105, 15779–15784 (2008). Article CAS PubMed PubMed Central Google Scholar * Neef, A.B. & Luedtke, N.W. Dynamic
metabolic labeling of DNA _in vivo_ with arabinosyl nucleosides. _Proc. Natl. Acad. Sci. USA_ 108, 20404–20409 (2011). Article CAS PubMed PubMed Central Google Scholar * Guan, L., van
der Heijden, G.W., Bortvin, A. & Greenberg, M.M. Intracellular detection of cytosine incorporation in genomic DNA by using 5-ethynyl-2′-deoxycytidine. _ChemBioChem_ 12, 2184–2190 (2011).
Article CAS PubMed PubMed Central Google Scholar * Grammel, M., Hang, H. & Conrad, N.K. Chemical reporters for monitoring RNA synthesis and poly(A) tail dynamics. _ChemBioChem_ 13,
1112–1115 (2012). Article CAS PubMed PubMed Central Google Scholar * Yamakoshi, H. et al. Imaging of EdU, an alkyne-tagged cell proliferation probe, by Raman microscopy. _J. Am. Chem.
Soc._ 27, 6102–6105 (2011). THIS STUDY DESCRIBES THE DIRECT IMAGING OF ALKYNE-TAGGED DNA BY RAMAN SPECTROSCOPY. Article CAS Google Scholar * Song, C.X. et al. Selective chemical labeling
reveals the genome-wide distribution of 5-hydroxymethylcytosine. _Nat. Biotechnol._ 29, 68–72 (2011). THIS STUDY DESCRIBES A CHEMOENZYMATIC METHOD FOR BIOORTHOGONAL DETECTION AND ENRICHMENT
OF 5-HMC MODIFICATION OF NUCLEIC ACIDS. Article CAS PubMed Google Scholar * He, Y.F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA.
_Science_ 333, 1303–1307 (2011). Article CAS PubMed PubMed Central Google Scholar * Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.
_Science_ 333, 1300–1303 (2011). Article CAS PubMed PubMed Central Google Scholar * Liu, C.C. & Schultz, P.G. Adding new chemistries to the genetic code. _Annu. Rev. Biochem._ 79,
413–444 (2010). Article CAS PubMed Google Scholar * Greiss, S. & Chin, J.W. Expanding the genetic code of an animal. _J. Am. Chem. Soc._ 133, 14196–14199 (2011). Article CAS PubMed
PubMed Central Google Scholar * Bianco, A., Townsley, F.M., Greiss, S., Lang, K. & Chin, J.W. Expanding the genetic code of _Drosophila melanogaster_. _Nat. Chem. Biol._ 8, 748–750
(2012). Article CAS PubMed Google Scholar * Lang, K. et al. Genetic encoding of bicyclononynes and _trans_-cyclooctenes for site-specific protein labeling _in vitro_ and in live
mammalian cells via rapid fluorogenic Diels-Alder reactions. _J. Am. Chem. Soc._ 134, 10317–10320 (2012). THIS STUDY DESCRIBES THE RAPID AND SELECTIVE BIOORTHOGONAL REACTION IN CELLS USING
SITE-SPECIFIC NONCANONICAL AMINO ACID INCORPORATION AND TETRAZINE REAGENTS. Article CAS PubMed PubMed Central Google Scholar * Johnson, J.A., Lu, Y.Y., Van Deventer, J.A. & Tirrell,
D.A. Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. _Curr. Opin. Chem. Biol._ 14, 774–780 (2010). Article CAS PubMed
PubMed Central Google Scholar * Liu, J., Xu, Y., Stoleru, D. & Salic, A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. _Proc. Natl. Acad. Sci. USA_
109, 413–418 (2012). Article CAS PubMed Google Scholar * Deal, R.B., Henikoff, J.G. & Henikoff, S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of
histones. _Science_ 328, 1161–1164 (2010). Article CAS PubMed PubMed Central Google Scholar * Eichelbaum, K., Winter, M., Diaz, M.B., Herzig, S. & Krijgsveld, J. Selective
enrichment of newly synthesized proteins for quantitative secretome analysis. _Nat. Biotechnol._ 30, 984–990 (2012). Article CAS PubMed Google Scholar * Howden, A.J. et al. QuaNCAT:
quantitating proteome dynamics in primary cells. _Nat. Methods_ 10, 343–346 (2013). Article CAS PubMed PubMed Central Google Scholar * Dieterich, D.C. et al. _In situ_ visualization and
dynamics of newly synthesized proteins in rat hippocampal neurons. _Nat. Neurosci._ 13, 897–905 (2010). Article CAS PubMed PubMed Central Google Scholar * Tcherkezian, J., Brittis,
P.A., Thomas, F., Roux, P.P. & Flanagan, J.G. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. _Cell_ 141, 632–644 (2010). Article CAS
PubMed PubMed Central Google Scholar * Ngo, J.T. et al. Cell-selective metabolic labeling of proteins. _Nat. Chem. Biol._ 5, 715–717 (2009). Article CAS PubMed PubMed Central Google
Scholar * Grammel, M., Zhang, M.M. & Hang, H.C. Orthogonal alkynyl amino acid reporter for selective labeling of bacterial proteomes during infection. _Angew. Chem. Int. Ed. Engl._ 49,
5970–5974 (2010). Article CAS PubMed PubMed Central Google Scholar * Grammel, M., Dossa, P.D., Taylor-Salmon, E. & Hang, H.C. Cell-selective labeling of bacterial proteomes with an
orthogonal phenylalanine amino acid reporter. _Chem. Commun. (Camb.)_ 48, 1473–1474 (2012). Article CAS Google Scholar * Saxon, E. & Bertozzi, C.R. Cell surface engineering by a
modified Staudinger reaction. _Science_ 287, 2007–2010 (2000). THIS STUDY DESCRIBES THE STAUNDINGER LIGATION REACTION BETWEEN FUNCTIONALIZED TRIARYLPHOSPHINE REAGENTS AND CELL-SURFACE
GLYCANS THAT WERE METABOLICALLY LABELED WITH AZIDE-MODIFIED MONOSACCHARIDE REPORTER. AS TRIARYLPHOSPHINE AND ALKYL AZIDES ARE BOTH ABIOTIC, THIS STUDY IS THE FIRST EXAMPLE OF A
'BIOORTHOGONAL' LIGATION REACTION. CAS PubMed Google Scholar * Laughlin, S.T. & Bertozzi, C.R. Imaging the glycome. _Proc. Natl. Acad. Sci. USA_ 106, 12–17 (2009). Article
CAS PubMed Google Scholar * Yarema, K.J., Goon, S. & Bertozzi, C.R. Metabolic selection of glycosylation defects in human cells. _Nat. Biotechnol._ 19, 553–558 (2001). THIS STUDY
HIGHLIGHTS THE APPLICATION OF CHEMICAL REPORTER LABELING AND SELECTION FOR IDENTIFICATION OF GENETIC MUTATIONS INVOLVED IN HUMAN DISEASE. Article CAS PubMed Google Scholar * Laughlin,
S.T., Baskin, J.M., Amacher, S.L. & Bertozzi, C.R. _In vivo_ imaging of membrane-associated glycans in developing zebrafish. _Science_ 320, 664–667 (2008). THIS STUDY DESCRIBES DYNAMIC
BIOORTHOGONAL LABELING AND IMAGING IN WHOLE ANIMALS. Article CAS PubMed PubMed Central Google Scholar * Dumont, A., Malleron, A., Awwad, M., Dukan, S. & Vauzeilles, B.
Click-mediated labeling of bacterial membranes through metabolic modification of the lipopolysaccharide inner core. _Angew. Chem. Int. Ed. Engl._ 51, 3143–3146 (2012). Article CAS PubMed
Google Scholar * Liu, F., Aubry, A.J., Schoenhofen, I.C., Logan, S.M. & Tanner, M.E. The engineering of bacteria bearing azido-pseudaminic acid–modified flagella. _ChemBioChem_ 10,
1317–1320 (2009). Article CAS PubMed Google Scholar * Swarts, B.M. et al. Probing the mycobacterial trehalome with bioorthogonal chemistry. _J. Am. Chem. Soc._ 134, 16123–16126 (2012).
Article CAS PubMed PubMed Central Google Scholar * Anderson, C.T., Wallace, I.S. & Somerville, C.R. Metabolic click-labeling with a fucose analog reveals pectin delivery,
architecture, and dynamics in _Arabidopsis_ cell walls. _Proc. Natl. Acad. Sci. USA_ 109, 1329–1334 (2012). Article CAS PubMed PubMed Central Google Scholar * Hart, G.W., Slawson, C.,
Ramirez-Correa, G. & Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. _Annu. Rev. Biochem._ 80, 825–858
(2011). Article CAS PubMed PubMed Central Google Scholar * Vocadlo, D.J., Hang, H.C., Kim, E.J., Hanover, J.A. & Bertozzi, C.R. A chemical approach for identifying O-GlcNAc–modified
proteins in cells. _Proc. Natl. Acad. Sci. USA_ 100, 9116–9121 (2003). Article CAS PubMed PubMed Central Google Scholar * Zaro, B.W., Yang, Y.Y., Hang, H.C. & Pratt, M.R. Chemical
reporters for fluorescent detection and identification of O-GlcNAc–modified proteins reveal glycosylation of the ubiquitin ligase NEDD4–1. _Proc. Natl. Acad. Sci. USA_ 108, 8146–8151 (2011).
THIS STUDY DESCRIBES A LARGE-SCALE PROTEOMIC ANALYSIS OF O-GLCNACYLATION USING CHEMICAL REPORTER LABELING. Article CAS PubMed PubMed Central Google Scholar * Rexach, J.E., Clark, P.M.
& Hsieh-Wilson, L.C. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. _Nat. Chem. Biol._ 4, 97–106 (2008). Article CAS PubMed PubMed Central Google Scholar
* Rexach, J.E. et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. _Nat. Chem. Biol._ 6, 645–651 (2010). Article CAS PubMed PubMed Central
Google Scholar * Rexach, J.E. et al. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. _Nat. Chem. Biol._ 8, 253–261 (2012). THIS STUDY DESCRIBES A
LARGE-SCALE CHEMOENZYMATIC PROTEOMIC ANALYSIS OF DYNAMIC O-GLCNACYLATION IN THE BRAIN. Article CAS PubMed PubMed Central Google Scholar * Pratt, M.R. et al. Deconvoluting the functions
of polypeptide N-α-acetylgalactosaminyltransferase family members by glycopeptide substrate profiling. _Chem. Biol._ 11, 1009–1016 (2004). Article CAS PubMed Google Scholar * Zheng, T.
et al. Tracking _N_-acetyllactosamine on cell-surface glycans _in vivo_. _Angew. Chem. Int. Ed. Engl._ 50, 4113–4118 (2011). Article CAS PubMed PubMed Central Google Scholar * Resh,
M.D. Trafficking and signaling by fatty-acylated and prenylated proteins. _Nat. Chem. Biol._ 2, 584–590 (2006). Article CAS PubMed Google Scholar * Hang, H.C., Wilson, J.P. &
Charron, G. Bioorthogonal chemical reporters for analyzing protein lipidation and lipid trafficking. _Acc. Chem. Res._ 44, 699–708 (2011). Article CAS PubMed PubMed Central Google
Scholar * Kho, Y. et al. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. _Proc. Natl. Acad. Sci. USA_ 101, 12479–12484 (2004). Article CAS PubMed
PubMed Central Google Scholar * Charron, G., Tsou, L.K., Maguire, W., Yount, J.S. & Hang, H.C. Alkynyl-farnesol reporters for detection of protein S-prenylation in cells. _Mol.
Biosyst._ 7, 67–73 (2011). Article CAS PubMed Google Scholar * DeGraw, A.J. et al. Evaluation of alkyne-modified isoprenoids as chemical reporters of protein prenylation. _Chem. Biol.
Drug Des._ 76, 460–471 (2010). Article CAS PubMed PubMed Central Google Scholar * Charron, G., Li, M., MacDonald, M. & Hang, H.C. Prenylome profiling reveals S-farnesylation is
crucial for membrane targeting and antiviral activity of ZAP long-isoform. _Proc. Natl. Acad. Sci. USA_ http://dx.doi.org/10.1073/pnas.1302564110 (2013). THIS STUDY DESCRIBES THE PROTEOMIC
ANALYSIS OF S-PRENYLATED PROTEINS IN MAMMALIAN CELLS USING CHEMICAL REPORTER LABELING AND THE DISCOVERY OF ZINC-FINGER ANTIVIRAL PROTEIN L (ZAPL) FARNESYLATION–DEPENDENT ANTIVIRAL ACTIVITY.
* Burnaevskiy, N. et al. Proteolytic elimination of N-myristoyl modifications by the _Shigella_ virulence factor IpaJ. _Nature_ 496, 106–109 (2013). Article CAS PubMed PubMed Central
Google Scholar * Fukata, Y. & Fukata, M. Protein palmitoylation in neuronal development and synaptic plasticity. _Nat. Rev. Neurosci._ 11, 161–175 (2010). Article CAS PubMed Google
Scholar * Martin, B.R. & Cravatt, B.F. Large-scale profiling of protein palmitoylation in mammalian cells. _Nat. Methods_ 6, 135–138 (2009). THIS STUDY DESCRIBES A LARGE-SCALE PROTEOMIC
ANALYSIS OF S-PALMITOYLATED PROTEINS IN T CELLS USING CHEMICAL REPORTER LABELING. Article CAS PubMed PubMed Central Google Scholar * Wilson, J.P., Raghavan, A.S., Yang, Y.Y., Charron,
G. & Hang, H.C. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. _Mol. Cell. Proteomics_ 10,
M110.001198 (2011). Article PubMed PubMed Central CAS Google Scholar * Yount, J.S. et al. Palmitoylome profiling reveals S-palmitoylation–dependent antiviral activity of IFITM3. _Nat.
Chem. Biol._ 6, 610–614 (2010). THIS STUDY DESCRIBES THE PROTEOMIC ANALYSIS OF S-PALMITOYLATED PROTEINS IN A DENDRITIC CELL LINE USING CHEMICAL REPORTER LABELING AND THE DISCOVERY THAT
S-PALMITOYLATION OF IFITM3 IS CRUCIAL FOR HOST RESISTANCE TO INFLUENZA VIRUS INFECTION. Article CAS PubMed PubMed Central Google Scholar * Li, Y., Martin, B.R., Cravatt, B.F. &
Hofmann, S.L. DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. _J. Biol. Chem._ 287, 523–530 (2012). Article CAS
PubMed Google Scholar * Zhang, M.M., Tsou, L.K., Charron, G., Raghavan, A.S. & Hang, H.C. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. _Proc. Natl. Acad.
Sci. USA_ 107, 8627–8632 (2010). THIS STUDY DESCRIBES DUAL CHEMICAL REPORTER LABELING FOR THE TANDEM DETECTION OF PROTEIN TURNOVER AND S-PALMITOYLATION DYNAMICS IN MAMMALIAN CELLS, WHICH
REVEALED ACCELERATED S-PALMITOYLATION IN ACTIVATED T CELLS. Article CAS PubMed PubMed Central Google Scholar * Martin, B.R., Wang, C., Adibekian, A., Tully, S.E. & Cravatt, B.F.
Global profiling of dynamic protein palmitoylation. _Nat. Methods_ 9, 84–89 (2012). THIS STUDY DESCRIBES LARGE-SCALE PROFILING OF DYNAMIC S-PALMITOYLATION IN T CELLS USING CHEMICAL REPORTER
LABELING WITH SILAC. Article CAS Google Scholar * Zhang, C.H., Wu, P.-Y.J., Kelly, F.D., Nurse, P. & Hang, H.C. Quantitative control protein S-palmitoylation regulates meiotic entry
in fission yeast. _PLoS Biol._ 11, e1001501 (2013). THIS STUDY DESCRIBES THE APPLICATION OF PALMITOYLATION REPORTER IN FISSION YEAST AND THE DISCOVERY THAT PROTEIN S-PALMITOYLATION IN
CELLULAR DIFFERENTIATION CAN BE QUANTITATIVELY REGULATED BY A SINGLE PALMITOYLTRANSFERASE. Article CAS PubMed PubMed Central Google Scholar * Ching, W., Hang, H.C. & Nusse, R.
Lipid-independent secretion of a _Drosophila_ Wnt protein. _J. Biol. Chem._ 283, 17092–17098 (2008). Article CAS PubMed PubMed Central Google Scholar * Heal, W.P. et al. Bioorthogonal
chemical tagging of protein cholesterylation in living cells. _Chem. Commun. (Camb.)_ 47, 4081–4083 (2011). Article CAS Google Scholar * Jiang, H. et al. SIRT6 regulates TNF-α secretion
through hydrolysis of long-chain fatty acyl lysine. _Nature_ 496, 110–113 (2013). Article CAS PubMed PubMed Central Google Scholar * Jao, C.Y., Roth, M., Welti, R. & Salic, A.
Metabolic labeling and direct imaging of choline phospholipids _in vivo_. _Proc. Natl. Acad. Sci. USA_ 106, 15332–15337 (2009). Article CAS PubMed PubMed Central Google Scholar * Yang,
J., Seckute, J., Cole, C.M. & Devaraj, N.K. Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. _Angew. Chem. Int. Ed. Engl._ 51, 7476–7479 (2012). Article
CAS PubMed PubMed Central Google Scholar * Milne, S.B. et al. Capture and release of alkyne-derivatized glycerophospholipids using cobalt chemistry. _Nat. Chem. Biol._ 6, 205–207
(2010). THIS STUDY DESCRIBES THE ANALYSIS OF LIPID METABOLISM IN MAMMALIAN CELLS USING CHEMICAL REPORTER LABELING AND COBALT-ALKYNE CHEMISTRY FOR THE CAPTURE AND RELEASE OF LABELED LIPIDS.
Article CAS PubMed PubMed Central Google Scholar * Thiele, C. et al. Tracing fatty acid metabolism by click chemistry. _ACS Chem. Biol._ 7, 2004–2011 (2012). Article CAS PubMed
Google Scholar * Rangan, K.J., Yang, Y.Y., Charron, G. & Hang, H.C. Rapid visualization and large-scale profiling of bacterial lipoproteins with chemical reporters. _J. Am. Chem. Soc._
132, 10628–10629 (2010). Article CAS PubMed PubMed Central Google Scholar * Babu, M.M. et al. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted
lipoproteins. _J. Bacteriol._ 188, 2761–2773 (2006). Article CAS PubMed PubMed Central Google Scholar * Lin, H., Su, X. & He, B. Protein lysine acylation and cysteine succination by
intermediates of energy metabolism. _ACS Chem. Biol._ 7, 947–960 (2012). Article CAS PubMed PubMed Central Google Scholar * Yang, Y.Y., Ascano, J.M. & Hang, H.C. Bioorthogonal
chemical reporters for monitoring protein acetylation. _J. Am. Chem. Soc._ 132, 3640–3641 (2010). THIS STUDY DESCRIBES CHEMICAL REPORTERS FOR PROTEIN ACETYLATION THAT FUNCTION _IN VITRO_ AND
IN MAMMALIAN CELLS. Article CAS PubMed PubMed Central Google Scholar * Yang, Y.Y., Grammel, M. & Hang, H.C. Identification of lysine acetyltransferase p300 substrates using
4-pentynoyl-coenzyme A and bioorthogonal proteomics. _Bioorg. Med. Chem. Lett._ 21, 4976–4979 (2011); erratum 21, 6613 (2011). Article CAS PubMed Google Scholar * Arnesen, T. et al.
Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. _Proc. Natl. Acad. Sci. USA_ 106, 8157–8162 (2009). Article
CAS PubMed PubMed Central Google Scholar * Mukherjee, S., Hao, Y.H. & Orth, K. A newly discovered post-translational modification—the acetylation of serine and threonine residues.
_Trends Biochem. Sci._ 32, 210–216 (2007). Article CAS PubMed Google Scholar * Bao, X., Zhao, Q., Yang, T., Fung, Y.M. & Li, X.D. A chemical probe for lysine malonylation. _Angew.
Chem. Int. Ed. Engl._ 52, 4883–4886 (2013). Article CAS PubMed Google Scholar * Greer, E.L. & Shi, Y. Histone methylation: a dynamic mark in health, disease and inheritance. _Nat.
Rev. Genet._ 13, 343–357 (2012). Article CAS PubMed PubMed Central Google Scholar * Dalhoff, C., Lukinavicius, G., Klimasauskas, S. & Weinhold, E. Direct transfer of extended groups
from synthetic cofactors by DNA methyltransferases. _Nat. Chem. Biol._ 2, 31–32 (2006). THIS STUDY DESCRIBES THE SAM CHEMICAL REPORTER THAT CAN BE EFFICIENTLY USED BY A DNA
METHYLTRANSFERASE. Article CAS PubMed Google Scholar * Peters, W. et al. Enzymatic site-specific functionalization of protein methyltransferase substrates with alkynes for click
labeling. _Angew. Chem. Int. Ed. Engl._ 49, 5170–5173 (2010). Article CAS PubMed Google Scholar * Islam, K., Zheng, W., Yu, H., Deng, H. & Luo, M. Expanding cofactor repertoire of
protein lysine methyltransferase for substrate labeling. _ACS Chem. Biol._ 6, 679–684 (2011). Article CAS PubMed PubMed Central Google Scholar * Wang, R., Zheng, W., Yu, H., Deng, H.
& Luo, M. Labeling substrates of protein arginine methyltransferase with engineered enzymes and matched _S_-adenosyl-L-methionine analogues. _J. Am. Chem. Soc._ 133, 7648–7651 (2011).
Article CAS PubMed PubMed Central Google Scholar * Wang, R. et al. Profiling genome-wide chromatin methylation with engineered posttranslation apparatus within living cells. _J. Am.
Chem. Soc._ 135, 1048–1056 (2013). THIS STUDY DESCRIBES THE APPLICATION OF A PROTEIN METHYLATION REPORTER IN LIVING CELLS. Article CAS PubMed PubMed Central Google Scholar * Willnow,
S., Martin, M., Luscher, B. & Weinhold, E. A selenium-based click AdoMet analogue for versatile substrate labeling with wild-type protein methyltransferases. _ChemBioChem_ 13, 1167–1173
(2012). Article CAS PubMed Google Scholar * Bothwell, I.R. et al. Se-adenosyl-L-selenomethionine cofactor analogue as a reporter of protein methylation. _J. Am. Chem. Soc._ 134,
14905–14912 (2012). Article CAS PubMed PubMed Central Google Scholar * Gibson, B.A. & Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and
PARPs. _Nat. Rev. Mol. Cell Biol._ 13, 411–424 (2012). Article CAS PubMed Google Scholar * Jiang, H., Kim, J.H., Frizzell, K.M., Kraus, W.L. & Lin, H. Clickable NAD analogues for
labeling substrate proteins of poly(ADP-ribose) polymerases. _J. Am. Chem. Soc._ 132, 9363–9372 (2010). THIS STUDY DESCRIBES _IN VITRO_ CHEMICAL REPORTERS FOR PROTEIN ADP-RIBOSYLATION.
Article CAS PubMed PubMed Central Google Scholar * Shogren-Knaak, M.A., Alaimo, P.J. & Shokat, K.M. Recent advances in chemical approaches to the study of biological systems. _Annu.
Rev. Cell Dev. Biol._ 17, 405–433 (2001). Article CAS PubMed Google Scholar * Yarbrough, M.L. et al. AMPylation of Rho GTPases by _Vibrio_ VopS disrupts effector binding and downstream
signaling. _Science_ 323, 269–272 (2009). Article CAS PubMed Google Scholar * Woolery, A.R., Luong, P., Broberg, C.A. & Orth, K. AMPylation: something old is new again. _Front
Microbiol._ 1, 113 (2010). Article CAS PubMed PubMed Central Google Scholar * Müller, M.P. et al. The _Legionella_ effector protein DrrA AMPylates the membrane traffic regulator Rab1b.
_Science_ 329, 946–949 (2010). Article PubMed Google Scholar * Tan, Y. & Luo, Z.Q. _Legionella pneumophila_ SidD is a deAMPylase that modifies Rab1. _Nature_ 475, 506–509 (2011).
Article CAS PubMed PubMed Central Google Scholar * Grammel, M., Luong, P., Orth, K. & Hang, H.C. A chemical reporter for protein AMPylation. _J. Am. Chem. Soc._ 133, 17103–17105
(2011). THIS STUDY DESCRIBES A CHEMICAL REPORTER FOR PROTEIN AMPYLATION. Article CAS PubMed PubMed Central Google Scholar * Macek, B., Mann, M. & Olsen, J.V. Global and
site-specific quantitative phosphoproteomics: principles and applications. _Annu. Rev. Pharmacol. Toxicol._ 49, 199–221 (2009). Article CAS PubMed Google Scholar * Allen, J.J. et al. A
semisynthetic epitope for kinase substrates. _Nat. Methods_ 4, 511–516 (2007). THIS STUDY DESCRIBES THE UNION OF A 'BUMPED' ATP ANALOG AND MUTANT KINASE PAIR WITH TWO-STEP
ALKYLATION AND ANTIBODY DETECTION OF SPECIFIC KINASE SUBSTRATES. Article CAS PubMed PubMed Central Google Scholar * Blethrow, J.D., Glavy, J.S., Morgan, D.O. & Shokat, K.M. Covalent
capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. _Proc. Natl. Acad. Sci. USA_ 105, 1442–1447 (2008). Article CAS PubMed PubMed Central Google Scholar *
Paulsen, C.E. et al. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. _Nat. Chem. Biol._ 8, 57–64 (2012). THIS STUDY DESCRIBES THE PROTEOMIC ANALYSIS OF
SULFENYLATION WITH AN ALKYNE CHEMICAL PROBE AND DISCOVERY OF OXIDATION-ENHANCED KINASE ACTIVITY. Article CAS Google Scholar * Blackman, M.L., Royzen, M. & Fox, J.M. Tetrazine
ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. _J. Am. Chem. Soc._ 130, 13518–13519 (2008). THE TETRAZINE LIGATION HAS AFFORDED A RAPID AND SELECTIVE
BIOORTHOGONAL LIGATION REACTION FOR LIVE-CELL IMAGING STUDIES. Article CAS PubMed PubMed Central Google Scholar * Devaraj, N.K. & Weissleder, R. Biomedical applications of tetrazine
cycloadditions. _Acc. Chem. Res._ 44, 816–827 (2011). THE TETRAZINE LIGATION HAS AFFORDED A RAPID AND SELECTIVE BIOORTHOGONAL LIGATION REACTION FOR LIVE-CELL IMAGING STUDIES. Article CAS
PubMed PubMed Central Google Scholar * Pezacki, J.P. et al. Chemical contrast for imaging living systems: molecular vibrations drive CARS microscopy. _Nat. Chem. Biol._ 7, 137–145 (2011).
Article CAS PubMed PubMed Central Google Scholar * Chang, P.V., Dube, D.H., Sletten, E.M. & Bertozzi, C.R. A strategy for the selective imaging of glycans using caged metabolic
precursors. _J. Am. Chem. Soc._ 132, 9516–9518 (2010). Article CAS PubMed PubMed Central Google Scholar * Xie, R., Hong, S., Feng, L., Rong, J. & Chen, X. Cell-selective metabolic
glycan labeling based on ligand-targeted liposomes. _J. Am. Chem. Soc._ 134, 9914–9917 (2012). Article CAS PubMed Google Scholar * Meldal, M. & Tornøe, C.W. Cu-catalyzed azide-alkyne
cycloaddition. _Chem. Rev._ 108, 2952–3015 (2008). STUDIES BY THE MELDAL AND SHARPLESS LABORATORIES DEMONSTRATED THAT AZIDE-ALKYNE CYCLOADDITIONS COULD BE ACCELERATED WITH CU( I ) CATALYSTS
FOR BIOORTHOGONAL LABELING APPLICATIONS. Article CAS PubMed Google Scholar * Kolb, H.C., Finn, M.G. & Sharpless, K.B. Click chemistry: diverse chemical function from a few good
reactions. _Angew. Chem. Int. Ed. Engl._ 40, 2004–2021 (2001). STUDIES BY THE MELDAL AND SHARPLESS LABORATORIES DEMONSTRATED AZIDE-ALKYNE CYCLOADDITIONS COULD BE ACCELERATED WITH CU( I )
CATALYSTS FOR BIOORTHOGONAL LABELING APPLICATIONS. Article CAS PubMed Google Scholar * Huisgen, R. 1,3-Dipolar cycloadditions. past and future. _Angew. Chem. Int. Ed. Engl._ 2, 565–598
(1963). EARLY STUDIES OF CYCLOADDITION REACTIONS BY HUISGEN AND COWORKERS PROVIDED THE FOUNDATION FOR FUTURE BIOORTHOGONAL REACTIONS. Article Google Scholar Download references
ACKNOWLEDGEMENTS We thank K. Rangan and N. Westcott for helpful comments on the manuscript. H.C.H. acknowledges support from Ellison Medical Foundation and US National Institutes of
Health–National Institute of General Medical Sciences (1R01GM087544). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Laboratory of Chemical Biology and Microbial Pathogenesis, The Rockefeller
University, New York, New York, USA Markus Grammel & Howard C Hang Authors * Markus Grammel View author publications You can also search for this author inPubMed Google Scholar * Howard
C Hang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.G. and H.C.H. wrote this review. CORRESPONDING AUTHOR Correspondence to Howard C
Hang. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Grammel, M., Hang, H. Chemical reporters for biological discovery. _Nat Chem Biol_ 9, 475–484 (2013). https://doi.org/10.1038/nchembio.1296 Download citation * Received: 30 July 2012 *
Accepted: 18 June 2013 * Published: 18 July 2013 * Issue Date: August 2013 * DOI: https://doi.org/10.1038/nchembio.1296 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative
