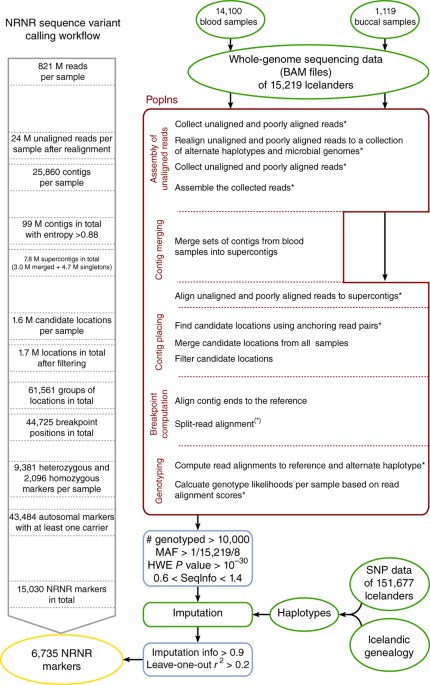
Diversity in non-repetitive human sequences not found in the reference genome
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Genomes usually contain some non-repetitive sequences that are missing from the reference genome and occur only in a population subset. Such non-repetitive, non-reference (NRNR)
sequences have remained largely unexplored in terms of their characterization and downstream analyses. Here we describe 3,791 breakpoint-resolved NRNR sequence variants called using PopIns
from whole-genome sequence data of 15,219 Icelanders. We found that over 95% of the 244 NRNR sequences that are 200 bp or longer are present in chimpanzees, indicating that they are
ancestral. Furthermore, 149 variant loci are in linkage disequilibrium (_r_2 > 0.8) with a genome-wide association study (GWAS) catalog marker, suggesting disease relevance. Additionally,
we report an association (_P_ = 3.8 × 10−8, odds ratio (OR) = 0.92) with myocardial infarction (23,360 cases, 300,771 controls) for a 766-bp NRNR sequence variant. Our results underline the
importance of including variation of all complexity levels when searching for variants that associate with disease. Access through your institution Buy or subscribe This is a preview of
subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value
online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more
Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS:
* Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS A LANDSCAPE OF COMPLEX TANDEM REPEATS WITHIN INDIVIDUAL
HUMAN GENOMES Article Open access 14 September 2023 A DRAFT HUMAN PANGENOME REFERENCE Article Open access 10 May 2023 A DEEP POPULATION REFERENCE PANEL OF TANDEM REPEAT VARIATION Article
Open access 23 October 2023 ACCESSION CODES PRIMARY ACCESSIONS NCBI REFERENCE SEQUENCE * KY503218 * KY508060 REFERENCES * Alkan, C., Coe, B.P. & Eichler, E.E. Genome structural variation
discovery and genotyping. _Nat. Rev. Genet._ 12, 363–376 (2011). Article CAS PubMed PubMed Central Google Scholar * Mills, R.E. et al. Mapping copy number variation by population-scale
genome sequencing. _Nature_ 470, 59–65 (2011). Article CAS PubMed PubMed Central Google Scholar * Kloosterman, W.P. et al. Chromothripsis as a mechanism driving complex _de novo_
structural rearrangements in the germline. _Hum. Mol. Genet._ 20, 1916–1924 (2011). Article CAS PubMed Google Scholar * Chaisson, M.J.P. et al. Resolving the complexity of the human
genome using single-molecule sequencing. _Nature_ 517, 608–611 (2014). Article PubMed PubMed Central Google Scholar * Hehir-Kwa, J.Y. et al. A high-quality human reference panel reveals
the complexity and distribution of genomic structural variants. _Nat. Commun._ 7, 12989 (2016). Article CAS PubMed PubMed Central Google Scholar * Telenti, A. et al. Deep sequencing of
10,000 human genomes. _Proc. Natl. Acad. Sci. USA_ 113, 11901–11906 (2016). Article CAS PubMed PubMed Central Google Scholar * Sudmant, P.H. et al. An integrated map of structural
variation in 2,504 human genomes. _Nature_ 526, 75–81 (2015). Article CAS PubMed PubMed Central Google Scholar * Mallick, S. et al. The Simons Genome Diversity Project: 300 genomes from
142 diverse populations. _Nature_ 538, 201–206 (2016). Article CAS PubMed PubMed Central Google Scholar * Kehr, B., Melsted, P. & Halldórsson, B.V. PopIns: population-scale
detection of novel sequence insertions. _Bioinformatics_ 32, 961–967 (2016). Article CAS PubMed Google Scholar * Schneider, V.A. et al. Evaluation of GRCh38 and _de novo_ haploid genome
assemblies demonstrates the enduring quality of the reference assembly. _bioRxiv_ http://dx.doi.org/10.1101/072116 (2016). * Gudbjartsson, D.F. et al. Sequence variants from whole genome
sequencing a large group of Icelanders. _Sci. Data_ 2, 150011 (2015). Article PubMed PubMed Central Google Scholar * Genovese, G. et al. Using population admixture to help complete maps
of the human genome. _Nat. Genet._ 45, 406–414 (2013). Article CAS PubMed Google Scholar * Kong, A. et al. A high-resolution map of the human genome. _Nat. Genet._ 31, 241–247 (2002).
Article CAS PubMed Google Scholar * International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. _Nature_ 409, 860–921 (2001). * Venter, C. et
al. The sequence of the human genome. _Science_ 291, 1304–1351 (2001). Article CAS PubMed Google Scholar * Abyzov, A. et al. Analysis of deletion breakpoints from 1,092 humans reveals
details of mutation mechanisms. _Nat. Commun._ 6, 7256 (2015). Article CAS PubMed Google Scholar * Levy, S. et al. The Diploid genome sequence of an individual human. _PLoS Biol._ 5,
e254 (2007). Article PubMed PubMed Central Google Scholar * Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP–trait associations. _Nucleic Acids Res._ 42, D1001–D1006
(2014). Article CAS PubMed Google Scholar * Olesen, M.S., Nielsen, M.W., Haunsø, S. & Svendsen, J.H. Atrial fibrillation: the role of common and rare genetic variants. _Eur. J. Hum.
Genet._ 22, 297–306 (2014). Article CAS PubMed Google Scholar * Osborne, T.F. Sterol regulatory element–binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin
action. _J. Biol. Chem._ 275, 32379–32382 (2000). Article CAS PubMed Google Scholar * Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for
coronary artery disease. _Nat. Genet._ 43, 333–338 (2011). Article CAS PubMed PubMed Central Google Scholar * Church, D.M. et al. Extending reference assembly models. _Genome Biol._ 16,
13 (2015). Article PubMed PubMed Central Google Scholar * Paten, B., Novak, A. & Haussler, D. Mapping to a reference genome structure. _arXiv_ https://arxiv.org/abs/1404.5010
(2014). * Eichler, E.E. et al. Missing heritability and strategies for finding the underlying causes of complex disease. _Nat. Rev. Genet._ 11, 446–450 (2010). Article CAS PubMed PubMed
Central Google Scholar * Manolio, T.A. et al. Finding the missing heritability of complex diseases. _Nature_ 461, 747–753 (2009). CAS PubMed PubMed Central Google Scholar * Kong, A. et
al. Fine-scale recombination rate differences between sexes, populations and individuals. _Nature_ 467, 1099–1103 (2010). Article CAS PubMed Google Scholar * Estrada, K. et al.
Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. _Nat. Genet._ 44, 491–501 (2012). Article CAS PubMed PubMed Central
Google Scholar * McMahon, F.J. et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. _Nat. Genet._ 42, 128–131 (2010). Article
CAS PubMed PubMed Central Google Scholar * arcOGEN Consortium & arcOGEN Collaborators. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide
association study. _Lancet_ 380, 815–823 (2012). * Manning, A.K. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits
and insulin resistance. _Nat. Genet._ 44, 659–669 (2012). Article CAS PubMed PubMed Central Google Scholar * Cai, Q. et al. Genome-wide association analysis in East Asians identifies
breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. _Nat. Genet._ 46, 886–890 (2014). Article CAS PubMed PubMed Central Google Scholar * Caporaso, N. et al. Genome-wide and
candidate gene association study of cigarette smoking behaviors. _PLoS One_ 4, e4653 (2009). Article PubMed PubMed Central Google Scholar * Wood, A.R. et al. Defining the role of common
variation in the genomic and biological architecture of adult human height. _Nat. Genet._ 46, 1173–1186 (2014). CAS PubMed PubMed Central Google Scholar * Shin, S.Y. et al. An atlas of
genetic influences on human blood metabolites. _Nat. Genet._ 46, 543–550 (2014). Article CAS PubMed PubMed Central Google Scholar * Trégouët, D.A. et al. Genome-wide haplotype
association study identifies the _SLC22A3_–_LPAL2_–_LPA_ gene cluster as a risk locus for coronary artery disease. _Nat. Genet._ 41, 283–285 (2009). Article PubMed Google Scholar * Perry,
J.R. et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. _Nature_ 514, 92–97 (2014). Article CAS PubMed PubMed Central Google Scholar *
Elks, C.E. et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. _Nat. Genet._ 42, 1077–1085 (2010). Article CAS PubMed PubMed
Central Google Scholar * Brown, C.C. et al. A genome-wide association analysis of temozolomide response using lymphoblastoid cell lines shows a clinically relevant association with _MGMT_.
_Pharmacogenet. Genomics_ 22, 796–802 (2012). Article CAS PubMed PubMed Central Google Scholar * Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological
pathways affect human height. _Nature_ 467, 832–838 (2010). Article CAS PubMed PubMed Central Google Scholar * Gudbjartsson, D.F. et al. Large-scale whole-genome sequencing of the
Icelandic population. _Nat. Genet._ 47, 435–444 (2015). Article CAS PubMed Google Scholar * Koressaar, T. & Remm, M. Enhancements and modifications of primer design program Primer3.
_Bioinformatics_ 23, 1289–1291 (2007). Article CAS PubMed Google Scholar * Untergasser, A. et al. Primer3—new capabilities and interfaces. _Nucleic Acids Res._ 40, e115 (2012). Article
CAS PubMed PubMed Central Google Scholar * Döring, A., Weese, D., Rausch, T. & Reinert, K. SeqAn an efficient, generic C++ library for sequence analysis. _BMC Bioinformatics_ 9, 11
(2008). Article PubMed PubMed Central Google Scholar * Kehr, B. et al. STELLAR: fast and exact local alignments. _BMC Bioinformatics_ 12, S15 (2011). Article PubMed PubMed Central
Google Scholar * Gu∂´ bjartsson, H. et al. GORpipe: a query tool for working with sequence data based on a Genomic Ordered Relational (GOR) architecture. _Bioinformatics_ 32, 3081–3088
(2016). Article Google Scholar * Styrkarsdottir, U. et al. Nonsense mutation in the _LGR4_ gene is associated with several human diseases and other traits. _Nature_ 497, 517–520 (2013).
Article CAS PubMed Google Scholar * Helgadottir, A. et al. Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. _Nat. Genet._
48, 634–639 (2016). Article CAS PubMed Google Scholar * Gretarsdottir, S. et al. A splice region variant in _LDLR_ lowers non–high density lipoprotein cholesterol and protects against
coronary artery disease. _PLoS Genet._ 11, e1005379 (2015). Article PubMed PubMed Central Google Scholar * Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of
insertions, deletions and gene fusions. _Genome Biol._ 14, R36 (2013). Article PubMed PubMed Central Google Scholar * Anders, S., Pyl, P.T. & Huber, W. HTSeq—a Python framework to
work with high-throughput sequencing data. _Bioinformatics_ 31, 166–169 (2015). Article CAS PubMed Google Scholar * Robinson, M.D. et al. A scaling normalization method for differential
expression analysis of RNA–seq data. _Genome Biol._ 11, R25 (2010). Article PubMed PubMed Central Google Scholar * Benson, D.A. et al. GenBank. _Nucleic Acids Res._ 45, D37–D42 (2017).
Article PubMed PubMed Central Google Scholar Download references AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * deCODE Genetics/Amgen, Inc., Reykjavik, Iceland Birte Kehr, Anna
Helgadottir, Pall Melsted, Hakon Jonsson, Hannes Helgason, Adalbjörg Jonasdottir, Aslaug Jonasdottir, Asgeir Sigurdsson, Arnaldur Gylfason, Gisli H Halldorsson, Snaedis Kristmundsdottir,
Hilma Holm, Unnur Thorsteinsdottir, Patrick Sulem, Agnar Helgason, Daniel F Gudbjartsson, Bjarni V Halldorsson & Kari Stefansson * Berlin Institute of Health, Berlin, Germany Birte Kehr
* School of Engineering and Natural Sciences, University of Iceland, Reykjavik, Iceland Pall Melsted, Hannes Helgason & Daniel F Gudbjartsson * Faculty of Medicine, School of Health
Sciences, University of Iceland, Reykjavik, Iceland Gudmundur Thorgeirsson, Unnur Thorsteinsdottir & Kari Stefansson * Department of Internal Medicine, Landspitali–National University
Hospital, Reykjavik, Iceland Gudmundur Thorgeirsson & Hilma Holm * Department of Clinical Biochemistry, Landspitali–National University Hospital, Reykjavik, Iceland Isleifur Olafsson *
Department of Anthropology, University of Iceland, Reykjavik, Iceland Agnar Helgason * School of Science and Engineering, Reykjavik University, Reykjavik, Iceland Bjarni V Halldorsson
Authors * Birte Kehr View author publications You can also search for this author inPubMed Google Scholar * Anna Helgadottir View author publications You can also search for this author
inPubMed Google Scholar * Pall Melsted View author publications You can also search for this author inPubMed Google Scholar * Hakon Jonsson View author publications You can also search for
this author inPubMed Google Scholar * Hannes Helgason View author publications You can also search for this author inPubMed Google Scholar * Adalbjörg Jonasdottir View author publications
You can also search for this author inPubMed Google Scholar * Aslaug Jonasdottir View author publications You can also search for this author inPubMed Google Scholar * Asgeir Sigurdsson View
author publications You can also search for this author inPubMed Google Scholar * Arnaldur Gylfason View author publications You can also search for this author inPubMed Google Scholar *
Gisli H Halldorsson View author publications You can also search for this author inPubMed Google Scholar * Snaedis Kristmundsdottir View author publications You can also search for this
author inPubMed Google Scholar * Gudmundur Thorgeirsson View author publications You can also search for this author inPubMed Google Scholar * Isleifur Olafsson View author publications You
can also search for this author inPubMed Google Scholar * Hilma Holm View author publications You can also search for this author inPubMed Google Scholar * Unnur Thorsteinsdottir View author
publications You can also search for this author inPubMed Google Scholar * Patrick Sulem View author publications You can also search for this author inPubMed Google Scholar * Agnar
Helgason View author publications You can also search for this author inPubMed Google Scholar * Daniel F Gudbjartsson View author publications You can also search for this author inPubMed
Google Scholar * Bjarni V Halldorsson View author publications You can also search for this author inPubMed Google Scholar * Kari Stefansson View author publications You can also search for
this author inPubMed Google Scholar CONTRIBUTIONS B.K., P.M., B.V.H. and K.S. designed the experiments. B.K., P.M., A.G., S.K., D.F.G. and B.V.H. implemented the methodology and analyzed the
call set. B.K., A. Helgadottir, H. Holm, P.S., D.F.G. and B.V.H. interpreted the association results. B.K., H. Helgason and G.H.H. analyzed gene expression. Aslaug Jonasdottir, Adalbjorg
Jonasdottir, and A.S. performed PCR verification and Sanger sequencing. U.T. oversaw the operations of the genotyping facilities. G.T., I.O., H. Holm and U.T. were responsible for phenotype
data acquisition. B.K. prepared tables and figures. B.K., H.J., A. Helgason and B.V.H. wrote the manuscript. All authors reviewed and approved the final manuscript. K.S. supervised the
study. CORRESPONDING AUTHORS Correspondence to Bjarni V Halldorsson or Kari Stefansson. ETHICS DECLARATIONS COMPETING INTERESTS B.K., A. Helgadottir, P.M., H.J., H. Helgason, Adalbjorg
Jonasdottir, Aslaug Jonasdottir, A.S., A.G., G.H.H., S.K., H. Holm, P.S., U.T., A. Helgason, D.F.G., B.V.H. and K.S. are all employees of deCODE Genetics/Amgen, Inc. INTEGRATED SUPPLEMENTARY
INFORMATION SUPPLEMENTARY FIGURE 1 PRIMER PAIRS DESIGNED FOR THE FIVE CATEGORIES OF NRNR SEQUENCE VARIANTS FOR VALIDATION BY SANGER SEQUENCING. For the categories “INS > 200” and “DEL
> INS”, two or three primer pairs were designed including at least one for each allele. For “INS < 200”, only a single primer pair was designed that may amplify in both alleles. For
“Different contig” always three and for “Singleton” always two primer pairs were designed. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figure 1, Supplementary
Tables 2, 7 and 8, and Supplementary Note (PDF 1950 kb) SUPPLEMENTARY DATA 1: NRNR SEQUENCES ANCHORED BY IMPUTED NRNR MARKERS. Sequences are given in FASTA format. (TXT 2129 kb)
SUPPLEMENTARY DATA 2: NRNR SEQUENCES ANCHORED BY FIXED NRNR MARKERS. Fixed are those markers that were predicted to have 100% frequency in Iceland. Sequences are given in FASTA format. (TXT
288 kb) SUPPLEMENTARY TABLE 1 List of imputed NRNR markers. (XLSX 623 kb) SUPPLEMENTARY TABLE 3 Details of Sanger sequencing validation experiments. (XLSX 65 kb) SUPPLEMENTARY TABLE 4 List
of fixed NRNR markers. (XLSX 48 kb) SUPPLEMENTARY TABLE 5 Overlap of NRNR markers with known variants and sequences. (XLSX 358 kb) SUPPLEMENTARY TABLE 6 Correlation with the GWAS catalog.
(XLSX 84 kb) SUPPLEMENTARY TABLE 9 Conversion to GenBank sequences. (XLSX 163 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kehr, B., Helgadottir,
A., Melsted, P. _et al._ Diversity in non-repetitive human sequences not found in the reference genome. _Nat Genet_ 49, 588–593 (2017). https://doi.org/10.1038/ng.3801 Download citation *
Received: 10 October 2016 * Accepted: 03 February 2017 * Published: 27 February 2017 * Issue Date: April 2017 * DOI: https://doi.org/10.1038/ng.3801 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative
