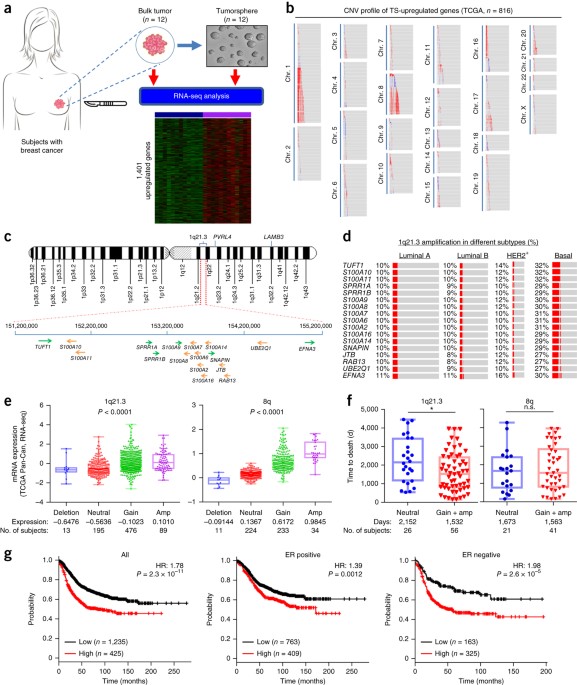
Chromosome 1q21. 3 amplification is a trackable biomarker and actionable target for breast cancer recurrence
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Tumor recurrence remains the main reason for breast cancer–associated mortality, and there are unmet clinical demands for the discovery of new biomarkers and development of
treatment solutions to benefit patients with breast cancer at high risk of recurrence. Here we report the identification of chromosomal copy-number amplification at 1q21.3 that is enriched
in subpopulations of breast cancer cells bearing characteristics of tumor-initiating cells (TICs) and that strongly associates with breast cancer recurrence. Amplification is present in
∼10–30% of primary tumors but in more than 70% of recurrent tumors, regardless of breast cancer subtype. Detection of amplification in cell-free DNA (cfDNA) from blood is strongly associated
with early relapse in patients with breast cancer and could also be used to track the emergence of tumor resistance to chemotherapy. We further show that 1q21.3-encoded S100 calcium-binding
protein (S100A) family members, mainly S100A7, S100A8, and S100A9 (S100A7/8/9), and IL-1 receptor–associated kinase 1 (IRAK1) establish a reciprocal feedback loop driving tumorsphere
growth. Notably, this functional circuitry can be disrupted by the small-molecule kinase inhibitor pacritinib, leading to preferential impairment of the growth of 1q21.3-amplified breast
tumors. Our study uncovers the 1q21.3-directed S100A7/8/9–IRAK1 feedback loop as a crucial component of breast cancer recurrence, serving as both a trackable biomarker and an actionable
therapeutic target for breast cancer. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through
your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this
journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now
Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS EXPLORING THE PROGNOSTIC SIGNIFICANCE OF ARM-LEVEL COPY NUMBER ALTERATIONS IN TRIPLE-NEGATIVE BREAST CANCER Article Open access 14 May 2024
_PIK3CA_ CO-OCCURRING MUTATIONS AND COPY-NUMBER GAIN IN HORMONE RECEPTOR POSITIVE AND HER2 NEGATIVE BREAST CANCER Article Open access 18 February 2022 COMPLEX REARRANGEMENTS FUEL ER+ AND
HER2+ BREAST TUMOURS Article Open access 08 January 2025 ACCESSION CODES PRIMARY ACCESSIONS GENE EXPRESSION OMNIBUS * GSE84054 REFERENCES * Al-Hajj, M., Wicha, M.S., Benito-Hernandez, A.,
Morrison, S.J. & Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. _Proc. Natl. Acad. Sci. USA_ 100, 3983–3988 (2003). Article CAS PubMed PubMed Central
Google Scholar * Nguyen, L.V., Vanner, R., Dirks, P. & Eaves, C.J. Cancer stem cells: an evolving concept. _Nat. Rev. Cancer_ 12, 133–143 (2012). Article CAS PubMed Google Scholar *
Mitra, A., Mishra, L. & Li, S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. _Oncotarget_ 6, 10697–10711 (2015). Article PubMed PubMed Central Google Scholar * Ginestier,
C. et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. _Cell Stem Cell_ 1, 555–567 (2007). Article CAS PubMed PubMed
Central Google Scholar * Marcato, P. et al. Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. _Mol. Oncol._ 9, 17–31 (2015). Article
CAS PubMed Google Scholar * Marcato, P. et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of
metastasis. _Stem Cells_ 29, 32–45 (2011). Article CAS PubMed Google Scholar * Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer
genomics data. _Cancer Discov._ 2, 401–404 (2012). Article PubMed Google Scholar * Ciriello, G. et al. Comprehensive molecular portraits of invasive lobular breast cancer. _Cell_ 163,
506–519 (2015). Article CAS PubMed PubMed Central Google Scholar * Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. _Sci.
Signal._ 6, pl1 (2013). Article CAS PubMed PubMed Central Google Scholar * Gyoőrffy, B., Surowiak, P., Budczies, J. & Lánczky, A. Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. _PLoS One_ 8, e82241 (2013). Article CAS Google Scholar * Pereira, B. et al. The somatic mutation
profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. _Nat. Commun._ 7, 11479 (2016). Article CAS PubMed PubMed Central Google Scholar * Curtis, C. et
al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. _Nature._ 486, 346–352 (2012). Article CAS PubMed PubMed Central Google Scholar *
Carreira, S. et al. Tumor clone dynamics in lethal prostate cancer. _Sci. Transl. Med._ 6, 254ra125 (2014). Article CAS PubMed PubMed Central Google Scholar * Gevensleben, H. et al.
Noninvasive detection of _HER2_ amplification with plasma DNA digital PCR. _Clin. Cancer Res._ 19, 3276–3284 (2013). Article CAS PubMed PubMed Central Google Scholar * Miotke, L., Lau,
B.T., Rumma, R.T. & Ji, H.P. High sensitivity detection and quantitation of DNA copy number and single nucleotide variants with single color droplet digital PCR. _Anal. Chem._ 86,
2618–2624 (2014). Article CAS PubMed PubMed Central Google Scholar * Pearson, A. et al. High-level clonal _FGFR_ amplification and response to FGFR inhibition in a translational
clinical trial. _Cancer Discov._ 6, 838–851 (2016). Article CAS PubMed PubMed Central Google Scholar * Romanel, A. et al. Plasma AR and abiraterone-resistant prostate cancer. _Sci.
Transl. Med._ 7, 312re10 (2015). Article CAS PubMed PubMed Central Google Scholar * Shoda, K. et al. Monitoring the _HER2_ copy number status in circulating tumor DNA by droplet digital
PCR in patients with gastric cancer. _Gastric Cancer_ 20, 126–135 (2016). Article CAS PubMed Google Scholar * Alix-Panabières, C. & Pantel, K. Clinical applications of circulating
tumor cells and circulating tumor DNA as liquid biopsy. _Cancer Discov._ 6, 479–491 (2016). Article CAS PubMed Google Scholar * Chan, D. et al. Phase II study of gemcitabine and
carboplatin in metastatic breast cancers with prior exposure to anthracyclines and taxanes. _Invest. New Drugs_ 28, 859–865 (2010). Article CAS PubMed Google Scholar * Wang, X. et al.
S100A14, a mediator of epithelial–mesenchymal transition, regulates proliferation, migration and invasion of human cervical cancer cells. _Am. J. Cancer Res._ 5, 1484–1495 (2015). CAS
PubMed PubMed Central Google Scholar * Tanaka, M. et al. Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion.
_BMC Cancer_ 15, 53 (2015). Article CAS PubMed PubMed Central Google Scholar * Bresnick, A.R., Weber, D.J. & Zimmer, D.B. S100 proteins in cancer. _Nat. Rev. Cancer_ 15, 96–109
(2015). Article CAS PubMed PubMed Central Google Scholar * Zhang, Q. et al. Downregulation of 425G>A variant of calcium-binding protein S100A14 associated with poor
differentiation and prognosis in gastric cancer. _J. Cancer Res. Clin. Oncol._ 141, 691–703 (2015). Article CAS PubMed Google Scholar * Cho, H. et al. The role of S100A14 in epithelial
ovarian tumors. _Oncotarget_ 5, 3482–3496 (2014). PubMed PubMed Central Google Scholar * Wee, Z.N. et al. IRAK1 is a therapeutic target that drives breast cancer metastasis and resistance
to paclitaxel. _Nat. Commun._ 6, 8746 (2015). Article CAS PubMed Google Scholar * Beauverd, Y., McLornan, D.P. & Harrison, C.N. Pacritinib: a new agent for the management of
myelofibrosis?. _Expert Opin. Pharmacother._ 16, 2381–2390 (2015). Article CAS PubMed Google Scholar * Singer, J.W. et al. Comprehensive kinase profile of pacritinib, a
nonmyelosuppressive Janus kinase 2 inhibitor. _J. Exp. Pharmacol._ 16, 11–19 (2016). Article Google Scholar * Phase 1/2 study of pacritinib, a next generation JAK2/FLT3 inhibitor, in
myelofibrosis or other myeloid malignancies. _Hematol. Oncol._ 9, 137 (2016). * Crowley, E., Di Nicolantonio, F., Loupakis, F. & Bardelli, A. Liquid biopsy: monitoring cancer-genetics in
the blood. _Nat. Rev. Clin. Oncol._ 10, 472–484 (2013). Article CAS PubMed Google Scholar * Siravegna, G. & Bardelli, A. Genotyping cell-free tumor DNA in the blood to detect
residual disease and drug resistance. _Genome Biol._ 15, 449 (2014). Article PubMed PubMed Central Google Scholar * Dawson, S.J. et al. Analysis of circulating tumor DNA to monitor
metastatic breast cancer. _N. Engl. J. Med._ 368, 1199–1209 (2013). Article CAS PubMed Google Scholar * Thress, K.S. et al. Acquired _EGFR_ C797S mutation mediates resistance to AZD9291
in non–small cell lung cancer harboring _EGFR_ T790M. _Nat. Med._ 21, 560–562 (2015). Article CAS PubMed PubMed Central Google Scholar * Siravegna, G. et al. Clonal evolution and
resistance to EGFR blockade in the blood of colorectal cancer patients. _Nat. Med._ 21, 795–801 (2015). Article CAS PubMed PubMed Central Google Scholar * Garcia-Murillas, I. et al.
Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. _Sci. Transl. Med._ 7, 302ra133 (2015). Article PubMed Google Scholar * Cancer Genome Atlas Network.
Comprehensive molecular portraits of human breast tumours. _Nature_ 490, 61–70 (2012). * Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour heterogeneity: a looking glass for cancer?
_Nat. Rev. Cancer_ 12, 323–334 (2012). Article CAS PubMed Google Scholar * Yates, L.R. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. _Nat.
Med._ 21, 751–759 (2015). Article CAS PubMed PubMed Central Google Scholar * Forshew, T. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep
sequencing of plasma DNA. _Sci. Transl. Med._ 4, 136ra68 (2012). Article CAS PubMed Google Scholar * Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by
sequencing of plasma DNA. _Nature_ 497, 108–112 (2013). Article CAS PubMed Google Scholar * Beaver, J.A. et al. Detection of cancer DNA in plasma of patients with early-stage breast
cancer. _Clin. Cancer Res._ 20, 2643–2650 (2014). Article CAS PubMed PubMed Central Google Scholar * Duffy, M.J., Shering, S., Sherry, F., McDermott, E. & O'Higgins, N. CA
15-3: a prognostic marker in breast cancer. _Int. J. Biol. Markers_ 15, 330–333 (2000). Article CAS PubMed Google Scholar * Harris, L. et al. American Society of Clinical Oncology 2007
update of recommendations for the use of tumor markers in breast cancer. _J. Clin. Oncol._ 25, 5287–5312 (2007). Article CAS PubMed Google Scholar * Sturgeon, C. Practice guidelines for
tumor marker use in the clinic. _Clin. Chem._ 48, 1151–1159 (2002). CAS PubMed Google Scholar * Mermel, C.H. et al. GISTIC2.0 facilitates sensitive and confident localization of the
targets of focal somatic copy-number alteration in human cancers. _Genome Biol._ 12, R41 (2011). Article CAS PubMed PubMed Central Google Scholar * Ono, S. et al. A direct plasma assay
of circulating microRNA-210 of hypoxia can identify early systemic metastasis recurrence in melanoma patients. _Oncotarget_ 6, 7053–7064 (2015). Article PubMed PubMed Central Google
Scholar * McShane, L.M. et al. Reporting recommendations for tumor marker prognostic studies (REMARK). _J. Natl. Cancer Inst._ 97, 1180–1184 (2005). Article CAS PubMed Google Scholar
Download references ACKNOWLEDGEMENTS This work was supported by the Agency for Science, Technology and Research of Singapore (A*STAR), the Margie Petersen Breast Cancer Program, and the
Association of Breast Cancer (D.S.B.H.), Singapore Ministry of Health's National Medical Research Council (NMRC) Clinician Scientist Individual Research Grants (NMRC/CIRG/1464/2016 and
NMRC/CG/017/2013 to E.Y.T.; NMRC/CSA/015/2009 and NMRC/CSA-SI/0004/2015 to S.C.L.), and the Danish Cancer Society (R99-A6362 and R146-A9164 to H.J.D.). This work was also supported by an
A*STAR Graduate Academy (A*GA) SINGA (Singapore International Graduate Award) scholarship to G.O. We thank the Histopathology Department from the Institute of Molecular and Cell Biology,
A*STAR, for their service in IHC staining and analysis. AUTHOR INFORMATION Author notes * Jian Yuan Goh, Min Feng and Wenyu Wang: These authors contributed equally to this work. AUTHORS AND
AFFILIATIONS * Genome Institute of Singapore, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore Jian Yuan Goh, Min Feng, Wenyu Wang, Gokce Oguz, Siti Maryam J M
Yatim, Puay Leng Lee, Yi Bao, Wai Leong Tam, Suman Sarma, Alexander Lezhava & Qiang Yu * Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore,
Singapore Gokce Oguz & Qiang Yu * Department of Pathology, Cytogenetics Laboratory, Singapore General Hospital, Singapore Tse Hui Lim & Alvin S T Lim * Cancer Research Institute and
School of Pharmacy, Jinan University, Guangzhou, China Panpan Wang & Qiang Yu * Cancer Science Institute of Singapore, National University of Singapore, Singapore Wai Leong Tam & Soo
Chin Lee * Department of Oncology, Odense University Hospital, Odense, Denmark Annette R Kodahl & Henrik J Ditzel * Department of Cancer and Inflammation Research, Institute of
Molecular Medicine, University of Southern Denmark, Odense, Denmark Maria B Lyng & Henrik J Ditzel * Department of Translational Molecular Medicine, John Wayne Cancer Institute, Santa
Monica, California, USA Selena Y Lin & Dave S B Hoon * Division of Medical Oncology, National Cancer Centre Singapore, Singapore Yoon Sim Yap * Department of Haematology–Oncology,
National University Cancer Institute, National University Health System, Singapore Soo Chin Lee * Department of General Surgery, Tan Tock Seng Hospital, Singapore Ern Yu Tan * Institute of
Molecular and Cellular Biology, A*STAR, Biopolis, Singapore Ern Yu Tan * Cancer and Stem Cell Biology, Duke-NUS Medical School, Singapore Qiang Yu Authors * Jian Yuan Goh View author
publications You can also search for this author inPubMed Google Scholar * Min Feng View author publications You can also search for this author inPubMed Google Scholar * Wenyu Wang View
author publications You can also search for this author inPubMed Google Scholar * Gokce Oguz View author publications You can also search for this author inPubMed Google Scholar * Siti
Maryam J M Yatim View author publications You can also search for this author inPubMed Google Scholar * Puay Leng Lee View author publications You can also search for this author inPubMed
Google Scholar * Yi Bao View author publications You can also search for this author inPubMed Google Scholar * Tse Hui Lim View author publications You can also search for this author
inPubMed Google Scholar * Panpan Wang View author publications You can also search for this author inPubMed Google Scholar * Wai Leong Tam View author publications You can also search for
this author inPubMed Google Scholar * Annette R Kodahl View author publications You can also search for this author inPubMed Google Scholar * Maria B Lyng View author publications You can
also search for this author inPubMed Google Scholar * Suman Sarma View author publications You can also search for this author inPubMed Google Scholar * Selena Y Lin View author publications
You can also search for this author inPubMed Google Scholar * Alexander Lezhava View author publications You can also search for this author inPubMed Google Scholar * Yoon Sim Yap View
author publications You can also search for this author inPubMed Google Scholar * Alvin S T Lim View author publications You can also search for this author inPubMed Google Scholar * Dave S
B Hoon View author publications You can also search for this author inPubMed Google Scholar * Henrik J Ditzel View author publications You can also search for this author inPubMed Google
Scholar * Soo Chin Lee View author publications You can also search for this author inPubMed Google Scholar * Ern Yu Tan View author publications You can also search for this author inPubMed
Google Scholar * Qiang Yu View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Q.Y. supervised the project and contributed to the design and
interpretation of all experiments. J.Y.G. contributed to the design, conduct, and interpretation of all experiments. M.F. performed tumor sample preparation, genomic DNA and cfDNA
extraction, RNA-seq, gene knockdown, and tumorsphere assays. Y.B. performed overexpression and tumorsphere assays. W.W. and P.L.L. performed western blot analyses. G.O. performed
bioinformatics and statistical analyses. W.W., M.F., and S.M.J.M.Y. performed _in vivo_ experiments. T.H.L. and A.S.T.L. performed DNA FISH analysis. P.W., A.R.K., M.B.L., and S.Y.L.
contributed to collection of patient blood samples and clinical information. A.L. and S.S. contributed digital PCR analyses. W.L.T., Y.S.Y., D.S.B.H., H.J.D., S.C.L., and E.Y.T. provided
crucial reagents and patient samples and contributed to clinical data analyses and interpretation. J.Y.G. and Q.Y. wrote the manuscript with input from all co-authors. CORRESPONDING AUTHORS
Correspondence to Henrik J Ditzel, Soo Chin Lee, Ern Yu Tan or Qiang Yu. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures 1–11 and Supplementary Tables 1–15. (PDF 1596 kb) LIFE SCIENCES REPORTING SUMMARY (PDF 129 KB) SUPPLEMENTARY DATA Uncropped
Immunoblots. (PDF 754 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Goh, J., Feng, M., Wang, W. _et al._ Chromosome 1q21.3 amplification is a
trackable biomarker and actionable target for breast cancer recurrence. _Nat Med_ 23, 1319–1330 (2017). https://doi.org/10.1038/nm.4405 Download citation * Received: 26 May 2017 * Accepted:
18 August 2017 * Published: 25 September 2017 * Issue Date: 01 November 2017 * DOI: https://doi.org/10.1038/nm.4405 SHARE THIS ARTICLE Anyone you share the following link with will be able
to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative
