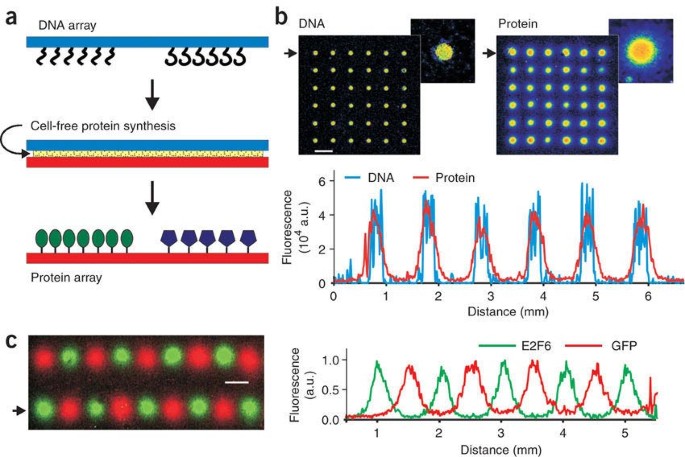
Printing protein arrays from DNA arrays
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

We describe a method, DNA array to protein array (DAPA), which allows the 'printing' of replicate protein arrays directly from a DNA array template using cell-free protein synthesis. At
least 20 copies of a protein array can be obtained from a single DNA array. DAPA eliminates the need for separate protein expression, purification and spotting, and also overcomes the
problem of long-term functional storage of surface-bound proteins.
We thank H. Liu for expert technical assistance, B. Korn (DKFZ, Heidelberg) and J. Taipele (University of Helsinki) for providing clones. Research at the Babraham Institute is supported by
the Biotechnology and Biological Sciences Research Council. Work on this project has been supported through the European Commission 6th Framework Programme Integrated Project MolTools.
Oda Stoevesandt, Elizabeth A Palmer, Farid Khan & Michael J Taussig
Present address: Present addresses: Protein Technology Group, Babraham Bioscience Technologies, Babraham Research Campus, Cambridge CB22 3AT, UK (O.S. and M.J.T.), MRC Virology Unit, Glasgow
G11 5JR, Scotland, UK (E.A.P), and University of Manchester, Manchester Interdisciplinary Biocentre, Manchester, M1 7DN, UK (F.K.).,
Mingyue He and Oda Stoevesandt: These authors contributed equally to this work.
Technology Research Group, The Babraham Institute, Cambridge, CB22 3AT, UK
Mingyue He, Oda Stoevesandt, Elizabeth A Palmer, Farid Khan & Michael J Taussig
Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, Uppsala, SE-75185, Sweden
M.H. and M.J.T. devised the principle of DAPA; M.H. developed and exemplified the method; O.S. optimized the method, devised the DAPA apparatus and designed and performed the experiments
shown in Figures 1 and 2; E.A.P. performed initial DAPA experiments, contributing examples to the data shown in Supplementary Figures 3 and 4; F.K. characterized the double-6His tag for
protein immobilization and protein detection methods; O.E. spotted the DNA template slides for experiments shown in Figure 1; M.H., O.S. and M.J.T. prepared the manuscript.
Supplementary Figures 1–4, Supplementary Methods (PDF 379 kb)
Anyone you share the following link with will be able to read this content:
