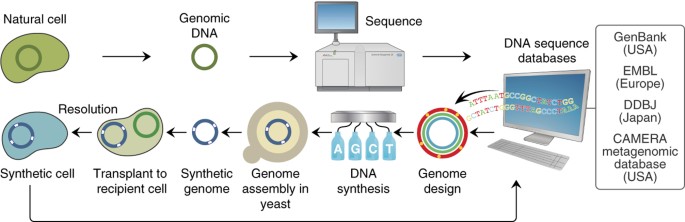
Programming biological operating systems: genome design, assembly and activation
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT The DNA technologies developed over the past 20 years for reading and writing the genetic code converged when the first synthetic cell was created 4 years ago. An outcome of this
work has been an extraordinary set of tools for synthesizing, assembling, engineering and transplanting whole bacterial genomes. Technical progress, options and applications for bacterial
genome design, assembly and activation are discussed. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS
Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS BUILDING GENOMES TO UNDERSTAND BIOLOGY Article Open access 02 December 2020 THE
TRANSCRIPTIONAL LANDSCAPE OF A REWRITTEN BACTERIAL GENOME REVEALS CONTROL ELEMENTS AND GENOME DESIGN PRINCIPLES Article Open access 24 May 2021 THE DESIGN AND ENGINEERING OF SYNTHETIC
GENOMES Article 06 November 2024 REFERENCES * Gibson, D.G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. _Science_ 329, 52–56 (2010). Article CAS
Google Scholar * Gibson, D.G. Oligonucleotide assembly in yeast to produce synthetic DNA fragments. _Methods Mol. Biol._ 852, 11–21 (2012). Article CAS Google Scholar * Gibson, D.G.
Enzymatic assembly of overlapping DNA fragments. _Methods Enzymol._ 498, 349–361 (2011). Article CAS Google Scholar * Gibson, D.G. Gene and genome construction in yeast. _Curr. Protoc.
Mol. Biol._ 94, 3.22 (2011). Google Scholar * Gibson, D.G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. _Nucleic Acids Res._ 37, 6984–6990
(2009). Article CAS Google Scholar * Gibson, D.G. et al. Complete chemical synthesis, assembly, and cloning of a _Mycoplasma genitalium_ genome. _Science_ 319, 1215–1220 (2008). Article
CAS Google Scholar * Gibson, D.G. et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic _Mycoplasma genitalium_ genome. _Proc. Natl. Acad. Sci.
USA_ 105, 20404–20409 (2008). Article CAS Google Scholar * Gibson, D.G., Smith, H.O., Hutchison, C.A. III., Venter, J.C. & Merryman, C. Chemical synthesis of the mouse mitochondrial
genome. _Nat. Methods_ 7, 901–903 (2010). Article CAS Google Scholar * Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. _Nat. Methods_ 6, 343–345
(2009). Article CAS Google Scholar * Benders, G.A. et al. Cloning whole bacterial genomes in yeast. _Nucleic Acids Res._ 38, 2558–2569 (2010). Article CAS Google Scholar * Lartigue, C.
et al. Genome transplantation in bacteria: changing one species to another. _Science_ 317, 632–638 (2007). Article CAS Google Scholar * Lartigue, C. et al. Creating bacterial strains
from genomes that have been cloned and engineered in yeast. _Science_ 325, 1693–1696 (2009). Article CAS Google Scholar * Venter, J.C. _Life at the Speed of Light: From the Double Helix
to the Dawn of Digital Life_ (Viking, 2013). * Ma, S., Tang, N. & Tian, J. DNA synthesis, assembly and applications in synthetic biology. _Curr. Opin. Chem. Biol._ 16, 260–267 (2012).
Article CAS Google Scholar * Ellis, T., Adie, T. & Baldwin, G.S. DNA assembly for synthetic biology: from parts to pathways and beyond. _Integr. Biol. (Camb.)_ 3, 109–118 (2011).
Article CAS Google Scholar * Itaya, M., Tsuge, K., Koizumi, M. & Fujita, K. Combining two genomes in one cell: stable cloning of the _Synechocystis_ PCC6803 genome in the _Bacillus
subtilis_ 168 genome. _Proc. Natl. Acad. Sci. USA_ 102, 15971–15976 (2005). Article CAS Google Scholar * Smailus, D.E., Warren, R.L. & Holt, R.A. Constructing large DNA segments by
iterative clone recombination. _Syst. Synth. Biol._ 1, 139–144 (2007). Article Google Scholar * Ma, H., Kunes, S., Schatz, P.J. & Botstein, D. Plasmid construction by homologous
recombination in yeast. _Gene_ 58, 201–216 (1987). Article CAS Google Scholar * Tagwerker, C. et al. Sequence analysis of a complete 1.66 Mb _Prochlorococcus marinus_ MED4 genome cloned
in yeast. _Nucleic Acids Res._ 40, 10375–10383 (2012). Article CAS Google Scholar * Karas, B.J., Tagwerker, C., Yonemoto, I.T., Hutchison, C.A. III. & Smith, H.O. Cloning the
_Acholeplasma laidlawii_ PG-8A genome in _Saccharomyces cerevisiae_ as a yeast centromeric plasmid. _ACS Synth. Biol._ 1, 22–28 (2012). Article CAS Google Scholar * Karas, B.J. et al.
Direct transfer of whole genomes from bacteria to yeast. _Nat. Methods_ 10, 410–412 (2013). Article CAS Google Scholar * Schwartz, D.C. & Cantor, C.R. Separation of yeast
chromosome-sized DNAs by pulsed field gradient gel electrophoresis. _Cell_ 37, 67–75 (1984). Article CAS Google Scholar * Noskov, V.N. et al. Assembly of large, high G+C bacterial DNA
fragments in yeast. _ACS Synth. Biol._ 1, 267–273 (2012). Article CAS Google Scholar * Kouprina, N. & Larionov, V. Selective isolation of genomic loci from complex genomes by
transformation-associated recombination cloning in the yeast _Saccharomyces cerevisiae_. _Nat. Protoc._ 3, 371–377 (2008). Article CAS Google Scholar * Kornberg, R.D. Eukaryotic
transcriptional control. _Trends Cell Biol._ 9, M46–M49 (1999). Article CAS Google Scholar * Kozak, M. Initiation of translation in prokaryotes and eukaryotes. _Gene_ 234, 187–208 (1999).
Article CAS Google Scholar * Marschall, P., Malik, N. & Larin, Z. Transfer of YACs up to 2.3 Mb intact into human cells with polyethylenimine. _Gene Ther._ 6, 1634–1637 (1999).
Article CAS Google Scholar * van Brabant, A.J., Buchanan, C.D., Charboneau, E., Fangman, W.L. & Brewer, B.J. An origin-deficient yeast artificial chromosome triggers a cell cycle
checkpoint. _Mol. Cell_ 7, 705–713 (2001). Article CAS Google Scholar * Carr, P.A. & Church, G.M. Genome engineering. _Nat. Biotechnol._ 27, 1151–1162 (2009). Article CAS Google
Scholar * Wang, H.H. et al. Programming cells by multiplex genome engineering and accelerated evolution. _Nature_ 460, 894–898 (2009). Article CAS Google Scholar * Isaacs, F.J. et al.
Precise manipulation of chromosomes _in vivo_ enables genome-wide codon replacement. _Science_ 333, 348–353 (2011). Article CAS Google Scholar * Ellis, H.M., Yu, D., DiTizio, T. &
Court, D.L. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. _Proc. Natl. Acad. Sci. USA_ 98, 6742–6746 (2001). Article CAS
Google Scholar * Dymond, J.S. et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. _Nature_ 477, 471–476 (2011). Article CAS Google Scholar *
Gaj, T., Gersbach, C.A. & Barbas, C.F. III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. _Trends Biotechnol._ 31, 397–405 (2013). Article CAS Google Scholar *
Gurdon, J.B. & Byrne, J.A. The first half-century of nuclear transplantation. _Proc. Natl. Acad. Sci. USA_ 100, 8048–8052 (2003). Article CAS Google Scholar * Gurdon, J.B. &
Wilmut, I. Nuclear transfer to eggs and oocytes. _Cold Spring Harb. Perspect. Biol._ 3, a002659 (2011). Article Google Scholar * Forster, A.C. & Church, G.M. Towards synthesis of a
minimal cell. _Mol. Syst. Biol._ 2, 45 (2006). Article Google Scholar * Forster, A.C. & Church, G.M. Synthetic biology projects _in vitro_. _Genome Res._ 17, 1–6 (2007). Article CAS
Google Scholar * Jewett, M.C. & Forster, A.C. Update on designing and building minimal cells. _Curr. Opin. Biotechnol._ 21, 697–703 (2010). Article CAS Google Scholar * Medema, M.H.,
van Raaphorst, R., Takano, E. & Breitling, R. Computational tools for the synthetic design of biochemical pathways. _Nat. Rev. Microbiol._ 10, 191–202 (2012). Article CAS Google
Scholar * Chan, L.Y., Kosuri, S. & Endy, D. Refactoring bacteriophage T7. _Mol. Syst. Biol._ 1, 2005.0018 (2005). Article Google Scholar * Jaschke, P.R., Lieberman, E.K., Rodriguez,
J., Sierra, A. & Endy, D. A fully decompressed synthetic bacteriophage oX174 genome assembled and archived in yeast. _Virology_ 434, 278–284 (2012). Article CAS Google Scholar *
Fraser, C.M. et al. The minimal gene complement of _Mycoplasma genitalium_. _Science_ 270, 397–403 (1995). Article CAS Google Scholar * Karr, J.R. et al. A whole-cell computational model
predicts phenotype from genotype. _Cell_ 150, 389–401 (2012). Article CAS Google Scholar * Dormitzer, P.R. et al. Synthetic generation of influenza vaccine viruses for rapid response to
pandemics. _Sci. Transl. Med._ 5, 185ra68 (2013). Article Google Scholar * Guye, P., Li, Y., Wroblewska, L., Duportet, X. & Weiss, R. Rapid, modular and reliable construction of
complex mammalian gene circuits. _Nucleic Acids Res._ 41, e156 (2013). Article Google Scholar * Temme, K., Zhao, D. & Voigt, C.A. Refactoring the nitrogen fixation gene cluster from
_Klebsiella oxytoca_. _Proc. Natl. Acad. Sci. USA_ 109, 7085–7090 (2012). Article Google Scholar * O'Neill, B.M. et al. An exogenous chloroplast genome for complex sequence
manipulation in algae. _Nucleic Acids Res._ 40, 2782–2792 (2012). Article CAS Google Scholar * Smith, H.O., Hutchison, C.A. III., Pfannkoch, C. & Venter, J.C. Generating a synthetic
genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. _Proc. Natl. Acad. Sci. USA_ 100, 15440–15445 (2003). Article CAS Google Scholar Download
references ACKNOWLEDGEMENTS I would like to thank past and present members of the synthetic biology groups at the J. Craig Venter Institute and Synthetic Genomics, Inc., for making all of
these technologies I discuss here possible, especially C. Venter, H. Smith and C. Hutchison, who had the vision to make a synthetic cell as early as 1995. I also thank M. LaPointe, T.
Richardson, J. Ward and J. Eads for their contributions to Figures 1, 2 and 4 and C. Hutchison for producing Figure 3. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * J. Craig Venter
Institute, La Jolla, California, USA Daniel G Gibson * Synthetic Genomics, Inc., La Jolla, California, USA Daniel G Gibson Authors * Daniel G Gibson View author publications You can also
search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Daniel G Gibson. ETHICS DECLARATIONS COMPETING INTERESTS D.G.G. is Vice President of DNA Technologies at
Synthetic Genomics, Inc., and holds employee stock shares in this company. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Gibson, D. Programming
biological operating systems: genome design, assembly and activation. _Nat Methods_ 11, 521–526 (2014). https://doi.org/10.1038/nmeth.2894 Download citation * Received: 11 December 2013 *
Accepted: 22 February 2014 * Published: 29 April 2014 * Issue Date: May 2014 * DOI: https://doi.org/10.1038/nmeth.2894 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative
