Autoimmune diseases — connecting risk alleles with molecular traits of the immune system
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

Autoimmune diseases affect approximately 8–9% of the world population. There are no cures and only limited therapies exist to treat symptoms.
Hundreds of genomic susceptibility loci have been discovered but, despite progress in fine-mapping causal genes and variant identification, the genetic mechanisms that trigger autoimmunity
are largely unknown.
Most risk variants that are likely to be causal are in non-coding sequences, particularly in regulatory regions known to be active in immune cell types; it is therefore crucial to understand
how enhancers and their targets affect gene regulation and immune function.
As a wider compendium of transcriptomic and epigenomic profiles for human immune cell types and cell states is built, we will be able to further pinpoint the conditions under which most risk
variants may be exerting their effect.
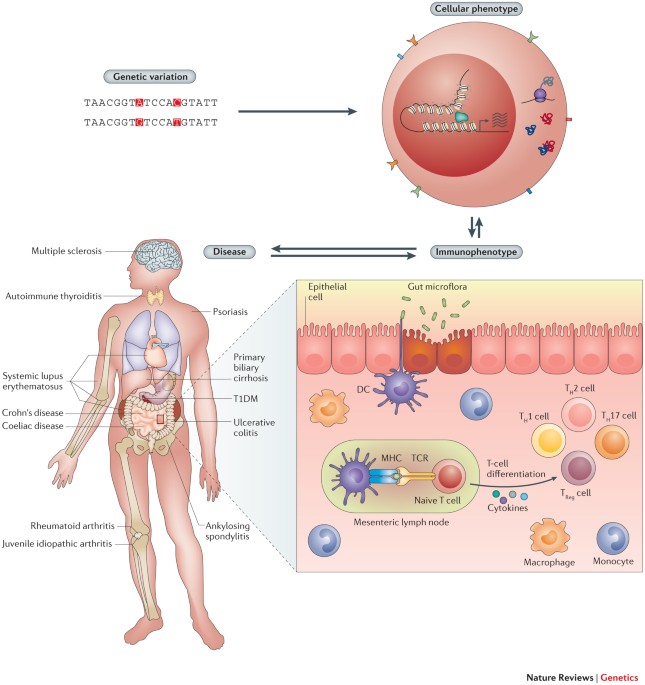
Profiling of immunophenotypes, such as signalling response, immune cell abundances and serum cytokine levels (among others), in thousands of individuals may help identify the key immune
functions and interactions involved in autoimmunity risk.
When the key immunophenotypes have been identified, longitudinal studies will be essential for revealing the causal relationships among intermediate phenotypes and elucidating the sequence
of events that lead to autoimmunity.
Genome-wide strategies have driven the discovery of more than 300 susceptibility loci for autoimmune diseases. However, for almost all loci, understanding of the mechanisms leading to
autoimmunity remains limited, and most variants that are likely to be causal are in non-coding regions of the genome. A critical next step will be to identify the in vivo and ex vivo
immunophenotypes that are affected by risk variants. To do this, key cell types and cell states that are implicated in autoimmune diseases will need to be defined. Functional genomic
annotations from these cell types and states can then be used to resolve candidate genes and causal variants. Together with longitudinal studies, this approach may yield pivotal insights
into how autoimmunity is triggered.
This work is supported in part by funding from the Swiss National Science Foundation (M.G.-A.), the US National Institutes of Health (1R01AR063759, 5U01GM092691-05, 1UH2AR067677-01 and U19
AI111224-01 (S.R.)) and the Doris Duke Charitable Foundation Grant #2013097 (S.R.). The authors thank the Raychaudhuri laboratory for their feedback, in particular H.-J. Westra and C. M.
Hong for their careful reading of the manuscript and C. Y. Fonseka for figure support.
Division of Genetics, and Division of Rheumatology, Department of Medicine, Immunology and Allergy, Brigham and Women's Hospital, Harvard Medical School, Boston, 02115, Massachusetts, USA
Program in Medical and Population Genetics, Broad Institute, Cambridge, 02142, Massachusetts, USA
Partners Center for Personalized Genetic Medicine, Boston, 02115, Massachusetts, USA
Center for Public Health Genomics, University of Virginia, Charlottesville, 22908, Virginia, USA
Faculty of Medical and Human Sciences, University of Manchester, Manchester, M13 9PL, UK
Department of Medicine, Karolinska Institutet and Karolinska University Hospital Solna, Stockholm, SE-171 77, Sweden
The part of the immune system that can react to specific pathogens and remember them to elicit a faster immune response if they are re-encountered.
When the immune system elicits an immune response against its own body (or non-pathogenic antigens that are commonly found in the body, such as commensal bacteria).
(T1DM). A disease characterized by autoreactivity to insulin-secreting β-cells found in the pancreas, resulting in high glucose levels in blood and urine.
(IBD). A term that groups together several conditions, such as Crohn's disease and ulcerative colitis, that are characterized by inflammation of the small intestine and colon. The
inflammation is thought to be caused by immune reaction to commensal bacteria.
(RA). Autoimmune disease causing joint inflammation and destruction of cartilage and bone. Patients with RA can present autoantibodies (seropositive) or not (seronegative).
(SLE). A disease characterized by autoreactivity to ubiquitous molecules, such as DNA or chromatin proteins, which can cause immune reactions against many parts of the body, some of the most
common being joints, skin, blood vessels, kidneys and nervous system.
(MS). A condition of autoimmunity against the myelin sheath that protects the central nervous system.
The mechanism by which the immune system does not attack its own body.
(GWAS). Studies in which genetic variants are ascertained across the whole genome in thousands of patients and thousands of controls, and statistical genetic associations (but not causality)
with disease are tested.
Quantitative measures of the strength of events. In genetic association studies, the odds ratio is often used as an effect size measure, as it reflects how many times more likely it is to
find the susceptibility variant in cases versus controls.
In this article, we define immunophenotypes as quantitative traits that reflect immune function. They include cell-type abundances, cell proliferation, signalling response, and cytokine and
chemokine serum protein levels.
A type of small cell-signalling protein that is secreted by some cells and affects the behaviour of others; cytokines are crucial participants in the immune response.
Inflammatory bowel disease presenting autoreactivity in any part of the gastrointestinal tract.
An autoimmune disease of the small intestine caused by an immune reaction to wheat.
A genetic association method that uses linkage disequilibrium and recombination principles in families to find markers linked to a disease locus (markers co-segregating with the disease
phenotype).
(HLA). A type of surface protein that presents antigens. HLA class I proteins present antigens from inside the cell and are expressed by most cells, whereas HLA class II proteins are
expressed only by antigen-presenting cells and present extracellular antigens to T cells.
(NF-κB). An important transcription factor involved in the immune response and B cell development.
A genetic association method that looks for alleles of a candidate gene (for example, based on a priori functional knowledge) that are associated with a disease, typically using a
case-control study design.
(LD). A phenomenon describing the nonrandom association (or co-segregation) between two alleles of different loci.
Groups of individuals pertaining to different ancestry populations.
Inference of alleles of non-genotyped variants based on nearby observed genotypes and using a reference panel from which the correlation structure between variants (or haplotypes) can be
learnt.
A concept referring to the heritability that has not been explained by known disease susceptibility loci (despite so many having been discovered with genome-wide association studies).
A technique that captures the protein-coding sequences of the genome (exons) and sequences them.
Distant transcriptional regulatory elements, often regulating expression in a cell-type- and cell-state-specific manner in cis, and often marked by specific chromatin modifications.
Several enhancer marks clustered together; super enhancers tend to be more cell-type specific than typical enhancer marks.
(eQTLs). Refers to a variant whose genotypes correlate with gene expression levels.
(Quantitative trait loci). Loci in the genome that are associated with a quantitative trait. For example, a single-nucleotide polymorphism associated with gene expression levels can be
called an expression QTL (eQTL).
A method for inferring causality of a trait (or modifiable exposure) on disease, taking advantage of the randomized trial nature of genetic variation (assuming a random mating pattern).
A phenomenon that occurs when one of the two alleles of a gene in an individual is expressed more than the other.
Anyone you share the following link with will be able to read this content:
