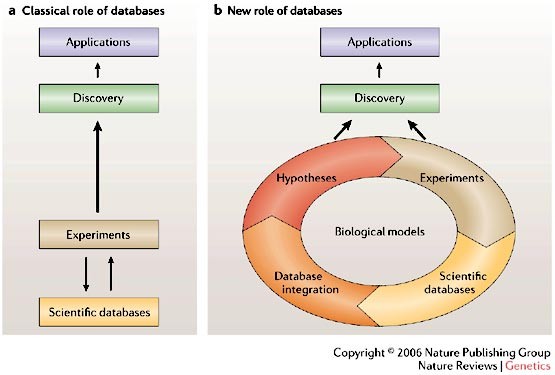
Addressing the problems with life-science databases for traditional uses and systems biology
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT A prerequisite to systems biology is the integration of heterogeneous experimental data, which are stored in numerous life-science databases. However, a wide range of obstacles that
relate to access, handling and integration impede the efficient use of the contents of these databases. Addressing these issues will not only be essential for progress in systems biology,
it will also be crucial for sustaining the more traditional uses of life-science databases. Access through your institution Buy or subscribe This is a preview of subscription content, access
via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy
this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: *
Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS JULIA FOR BIOLOGISTS Article 06 April 2023 RECONSTRUCTING
ORGANISMS IN SILICO: GENOME-SCALE MODELS AND THEIR EMERGING APPLICATIONS Article 21 September 2020 DIVERSIFYING THE CONCEPT OF MODEL ORGANISMS IN THE AGE OF -OMICS Article Open access 19
October 2023 REFERENCES * Kitano, H. Systems biology: a brief overview. _Science_ 295, 1662–1664 (2002). Article CAS PubMed Google Scholar * Pennisi, E. How will big pictures emerge from
a sea of biological data? _Science_ 309, 94 (2005). Article CAS PubMed Google Scholar * Roos, D. S. Computational biology. Bioinformatics — trying to swim in a sea of data. _Science_
291, 1260–1261 (2001). Article CAS PubMed Google Scholar * Augen, J. Information technology to the rescue! _Nature Biotechnol._ 19, BE39–BE40 (2001). Article CAS Google Scholar * Ge,
H., Walhout, A. J. & Vidal, M. Integrating 'omic' information: a bridge between genomics and systems biology. _Trends Genet._ 19, 551–560 (2003). Article CAS PubMed Google
Scholar * Carel, R. Practical data integration in biopharmaceutical research and development. _PharmaGenomics_ 22–35 (June 2003). * Galperin, M. Y. The Molecular Biology Database
Collection: 2006 update. _Nucleic Acids Res._ 34, D3–D5 (2006). Article CAS PubMed Google Scholar * Cerami, E. _Web services essentials_ (O'Reilly, Beijing; Sebastopol, California,
2002). Google Scholar * Sugawara, H. & Miyazaki, S. Biological SOAP servers and web services provided by the public sequence data bank. _Nucleic Acids Res._ 31, 3836–3839 (2003).
Article CAS PubMed PubMed Central Google Scholar * Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y. & Hattori, M. The KEGG resource for deciphering the genome. _Nucleic Acids Res._
32, D277–D280 (2004). Article CAS PubMed PubMed Central Google Scholar * Pillai, S. et al. SOAP-based services provided by the European Bioinformatics Institute. _Nucleic Acids Res._
33, W25–W28 (2005). Article CAS PubMed PubMed Central Google Scholar * Stajich, J. E. et al. The Bioperl toolkit: Perl modules for the life sciences. _Genome Res._ 12, 1611–1618 (2002).
Article CAS PubMed PubMed Central Google Scholar * Mangalam, H. The Bio * toolkits — a brief overview. _Brief. Bioinformatics_ 3, 296–302 (2002). Article PubMed Google Scholar *
Wang, L., Riethoven, J. J. & Robinson, A. XEMBL: distributing EMBL data in XML format. _Bioinformatics_ 18, 1147–1148 (2002). Article CAS PubMed Google Scholar * Bairoch, A. et al.
The Universal Protein Resource (UniProt). _Nucleic Acids Res._ 33, D154–D159 (2005). Article CAS PubMed Google Scholar * Luciano, J. S. PAX of mind for pathway researchers. _Drug Discov.
Today_ 10, 937–942 (2005). Article CAS PubMed Google Scholar * Lloyd, C. M., Halstead, M. D. & Nielsen, P. F. CellML: its future, present and past. _Prog. Biophys. Mol. Biol._ 85,
433–450 (2004). Article CAS PubMed Google Scholar * Spellman, P. T. et al. Design and implementation of microarray gene expression markup language (MAGE-ML). _Genome Biol._ 3,
RESEARCH0046 (2002). * Orchard, S. et al. Further steps in standardisation. Report of the second annual Proteomics Standards Initiative Spring Workshop (Siena, Italy 17–20th April 2005).
_Proteomics_ 5, 3552–3555 (2005). Article CAS PubMed Google Scholar * Hucka, M. et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical
network models. _Bioinformatics_ 19, 524–531 (2003). Article CAS PubMed Google Scholar * Green, M. L. & Karp, P. D. Genome annotation errors in pathway databases due to semantic
ambiguity in partial EC numbers. _Nucleic Acids Res._ 33, 4035–4039 (2005). Article CAS PubMed PubMed Central Google Scholar * Stevens, R. et al. TAMBIS: transparent access to multiple
bioinformatics information sources. _Bioinformatics_ 16, 184–185 (2000). Article CAS PubMed Google Scholar * Köhler, J., Philippi, S. & Lange, M. SEMEDA: ontology based semantic
integration of biological databases. _Bioinformatics_ 19, 2420–2427 (2003). Article PubMed Google Scholar * Ashburner, M. et al. Gene Ontology: tool for the unification of biology. The
Gene Ontology Consortium. _Nature Genet._ 25, 25–29 (2000). Article CAS PubMed Google Scholar * Philippi, S. & Köhler, J. Using XML technology for the ontology-based semantic
integration of life science databases. _IEEE Trans. Inf. Technol. Biomed._ 8, 154–160 (2004). Article PubMed Google Scholar * NC-IUBMB. _Enzyme Nomenclature 1992: Recommendations of the
Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes_ (Academic Press, San Diego, 1992). * Wheeler, D. L.
et al. Database resources of the National Center for Biotechnology Information: update. _Nucleic Acids Res._ 32, D35–D40 (2004). Article CAS PubMed PubMed Central Google Scholar *
Hendler, J. Communication. Science and the semantic web. _Science_ 299, 520–521 (2003). Article CAS PubMed Google Scholar * Noble, D. Will genomics revolutionise pharmaceutical R&D?
_Trends Biotechnol._ 21, 333–337 (2003). Article CAS PubMed Google Scholar * Smith, B., Köhler, J. & Kumar, A. On the application of formal principles to life science data: a case
study in the gene ontology. _Proc. Data Integr. Life Sci. First Int. Workshop_ 79–94 (2004). * Zhang, S. & Bodenreider, O. Law and order: assessing and enforcing compliance with
ontological modeling principles in the Foundational Model of Anatomy. _Comput. Biol. Med._ 6 Sep 2005 (doi:10.1016/j.compbiomed.2005.04.007). * van Helden, J. et al. Representing and
analysing molecular and cellular function using the computer. _Biol. Chem._ 381, 921–935 (2000). CAS PubMed Google Scholar * Bornberg-Bauer, E. & Paton, N. W. Conceptual data
modelling for bioinformatics. _Brief. Bioinformatics_ 3, 166–180 (2002). Article CAS PubMed Google Scholar * Nelson, M. R., Reisinger, S. J. & Henry, S. G. Designing databases to
store biological information. _BioSilico_ 1, 134–142 (2003). Article CAS Google Scholar * Taylor, C. F. et al. A systematic approach to modeling, capturing, and disseminating proteomics
experimental data. _Nature Biotechnol._ 21, 247–254 (2003). Article CAS Google Scholar * Ma, Z. & Chen, J. (eds) _Database Modeling in Biology: Practices and Challenges_ (Springer, in
the press). * Karp, P. D. et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. _Nucleic Acids Res._ 33, 6083–6089 (2005). Article CAS PubMed PubMed
Central Google Scholar * Searls, D. B. Data integration — connecting the dots. _Nature Biotechnol._ 21, 844–845 (2003). Article CAS Google Scholar * Karp, P. D. What we do not know
about sequence analysis and sequence databases. _Bioinformatics_ 14, 753–754 (1998). Article CAS PubMed Google Scholar * Camon, E. et al. The Gene Ontology Annotation (GOA) Database:
sharing knowledge in Uniprot with Gene Ontology. _Nucleic Acids Res._ 32, D262–D266 (2004). Article CAS PubMed PubMed Central Google Scholar * Gattiker, A. et al. Automated annotation
of microbial proteomes in SWISS-PROT. _Comput. Biol. Chem._ 27, 49–58 (2003). Article CAS PubMed Google Scholar * Garcia-Berthou, E. & Alcaraz, C. Incongruence between test
statistics and P values in medical papers. _BMC Med. Res. Methodol._ 4, 13 (2004). Article PubMed PubMed Central Google Scholar * Mecham, B. H. et al. Increased measurement accuracy for
sequence-verified microarray probes. _Physiol. Genomics_ 18, 308–315 (2004). Article CAS PubMed Google Scholar * Ntzani, E. E. & Ioannidis, J. P. Predictive ability of DNA
microarrays for cancer outcomes and correlates: an empirical assessment. _Lancet_ 362, 1439–1444 (2003). Article CAS PubMed Google Scholar * Hirschhorn, J. N., Lohmueller, K., Byrne, E.
& Hirschhorn, K. A comprehensive review of genetic association studies. _Genet. Med._ 4, 45–61 (2002). Article CAS PubMed Google Scholar * Müller, H., Naumann, F. & Freytag,
J.-C. Data quality in genome databases. _Proc. Conf. Inf. Qual. (IQ 03)_ 269–284 (2003). * Iliopoulos, I. et al. Evaluation of annotation strategies using an entire genome sequence.
_Bioinformatics_ 19, 717–726 (2003). Article CAS PubMed Google Scholar * Leser, U. & Hakenberg, J. What makes a gene name? Named entity recognition in the biomedical literature.
_Brief. Bioinformatics_ 6, 357–369 (2005). Article CAS PubMed Google Scholar * Resnik, D. B. Strengthening the United States' database protection laws: balancing public access and
private control. _Sci. Eng. Ethics_ 9, 301–318 (2003). Article PubMed Google Scholar * Maurer, S. M., Hugenholtz, P. B. & Onsrud, H. J. Intellectual property. Europe's database
experiment. _Science_ 294, 789–790 (2001). Article CAS PubMed Google Scholar * Merali, Z. & Giles, J. Databases in peril. _Nature_ 435, 1010–1011 (2005). Article CAS PubMed Google
Scholar * Ellis, L. B. & Kalumbi, D. The demise of public data on the web? _Nature Biotechnol._ 16, 1323–1324 (1998). Article CAS Google Scholar * Greenbaum, D. & Gerstein, M. A
universal legal framework as a prerequisite for database interoperability. _Nature Biotechnol._ 21, 979–982 (2003). Article CAS Google Scholar * Brazma, A. et al. Minimum information
about a microarray experiment (MIAME) — toward standards for microarray data. _Nature Genet._ 29, 365–371 (2001). Article CAS PubMed Google Scholar * Bourne, P. Will a biological
database be different from a biological journal? _PLoS Comput. Biol._ 1, 179–181 (2005). CAS PubMed Google Scholar * Berman, H. M. et al. The Protein Data Bank. _Nucleic Acids Res._ 28,
235–242 (2000). Article CAS PubMed PubMed Central Google Scholar * Rother, K. et al. Columba: multidimensional data integration of protein annotations. _Proc. Data Integr. Life Sci.
First Int. Workshop_ 156–171 (2004). * Zdobnov, E. M., Lopez, R., Apweiler, R. & Etzold, T. The EBI SRS server — recent developments. _Bioinformatics_ 18, 368–373 (2002). Article CAS
PubMed Google Scholar * Haas, L. M. et al. DiscoveryLink: a system for integrated access to life sciences data sources. _IBM Syst. J._ 40, 489–511 (2001). Article Google Scholar *
Köhler, J. et al. Linking experimental results, biological networks and sequence analysis methods using Ontologies and Generalised Data Structures. _In Silico Biol._ 5, 33–44 (2004). Google
Scholar * Stein, L. D. Integrating biological databases. _Nature Rev. Genet._ 4, 337–345 (2003). Article CAS PubMed Google Scholar * Köhler, J. Integration of life science databases.
_Drug Discov. Today_ 2, 61–69 (2004). Article Google Scholar * Matys, V. et al. TRANSFAC: transcriptional regulation, from patterns to profiles. _Nucleic Acids Res._ 31, 374–378 (2003).
Article CAS PubMed PubMed Central Google Scholar * Kolchanov, N. A. et al. Transcription Regulatory Regions Database (TRRD): its status in 2002. _Nucleic Acids Res._ 30, 312–317 (2002).
Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS The authors would like to thank C. Rawlings and P. Verrier for commenting on an earlier version of
this article. Furthermore we would like to thank the following individuals for exploring with us the pitfalls of life-science databases over the past years: J. Baumbach, J. Butz, E.
Kirchem, F. Klingert, S. Knop, B. Kormeier, I. Kupp, A. Neu, A. Rüegg, A. Skusa, B. Steuernagel, J. Taubert, P. Verrier and R. Winnenburg. S.P. gratefully acknowledges funding by the
European Science Foundation. Rothamsted Research receives grant-aided support from the UK Biotechnological and Biological Science Research Council. AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * Stephan Philippi is at the Department of Computer Science, University of Koblenz, PO Box 201602, Koblenz, 56016, Germany Stephan Philippi * Jacob Köhler is at the
Biomathematics and Bioinformatics Division, Rothamsted Research, Harpenden, AL5 2JQ, Hertfordshire, UK Jacob Köhler Authors * Stephan Philippi View author publications You can also search
for this author inPubMed Google Scholar * Jacob Köhler View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Stephan
Philippi. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. RELATED LINKS RELATED LINKS FURTHER INFORMATION BioJava BioPAX — Biological Pathways
Exchange BioPerl BioRuby CellML DiscoveryLink DNA Data Bank of Japan EC (enzyme class) numbers of the enzyme nomenclature Ensembl Trace Server European Bioinformatics Institute SRS server
European Bioinformatics Institute Extensible Markup Language (XML) Gene Ontology homepage Kyoto Encyclopedia of Genes and Genomes Microarray Gene Expression Data Society mySQL NCBI taxonomy
Nucleic Acids Research Database Categories List ONDEX Open Biomedical Ontologies Open Source Initiative License Index PostgreSQL Proteomics Standards Initiative — molecular interaction
Systems biology markup language Universal Protein Resource Web Services Activity GLOSSARY * Controlled vocabulary A standardized set of terms that can be used in a given application domain.
A prominent example is the enzyme class nomenclature, which describes classes of biochemical reaction. * Database management system A system that provides a means of storing, modifying and
extracting data from a database. * Evidence code A controlled vocabulary that is used to track the types of evidence that support a gene annotation. * Flat file Human readable,
non-standardized files that can be used to exchange the contents of life-science databases. * Ontology A commonly agreed definition of real-world concepts, such as 'protein' and
'enzyme', and their particular relationships, for example, an enzyme 'is a' protein. * Parser Software that reads a given input, such as a flat file, for further
processing. * Web service A standardized way to allow for interoperable machine-to-machine interaction over a network. * XML The extensible markup language (XML) is a standard for the
creation of application-specific, self-descriptive markup languages, which, for example, can be used for the definition of data-exchange formats. RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Philippi, S., Köhler, J. Addressing the problems with life-science databases for traditional uses and systems biology. _Nat Rev Genet_ 7,
482–488 (2006). https://doi.org/10.1038/nrg1872 Download citation * Published: 09 May 2006 * Issue Date: 01 June 2006 * DOI: https://doi.org/10.1038/nrg1872 SHARE THIS ARTICLE Anyone you
share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the
Springer Nature SharedIt content-sharing initiative
