Comprehensive phenotypic assessment of nonsense mutations in mitochondrial nd5 in mice
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Mitochondrial dysfunction induced by mitochondrial DNA (mtDNA) mutations has been implicated in various human diseases. A comprehensive analysis of mitochondrial genetic disorders
requires suitable animal models for human disease studies. While gene knockout via premature stop codons is a powerful method for investigating the unique functions of target genes,
achieving knockout of mtDNA has been rare. Here, we report the genotypes and phenotypes of heteroplasmic MT-ND5 gene-knockout mice. These mutant mice presented damaged mitochondrial cristae
in the cerebral cortex, hippocampal atrophy, and asymmetry, leading to learning and memory abnormalities. Moreover, mutant mice are susceptible to obesity and thermogenetic disorders. We
propose that these mtDNA gene-knockdown mice could serve as valuable animal models for studying the MT-ND5 gene and developing therapies for human mitochondrial disorders in the future.
SIMILAR CONTENT BEING VIEWED BY OTHERS MODELLING POLG MUTATIONS IN MICE UNRAVELS A CRITICAL ROLE OF POLΓΒ IN REGULATING PHENOTYPIC SEVERITY Article Open access 23 May 2025 A HIGH MUTATION
LOAD OF M.14597A>G IN _MT-ND6_ CAUSES LEIGH SYNDROME Article Open access 27 May 2021 ZEBRAFISH _POLG2_ KNOCK-OUT RECAPITULATES HUMAN POLG-DISORDERS; IMPLICATIONS FOR DRUG TREATMENT
Article Open access 20 April 2024 INTRODUCTION Mitochondria are essential organelles capable of producing chemical energy, and they play an important role in sustaining our survival. As
energy powerhouses, mitochondria supply >90% of the adenosine triphosphate (ATP) necessary for cellular metabolism1. Additionally, mitochondria play crucial roles in various cellular
processes, including the regulation of ion homeostasis, redox status, cell survival, and cell death2. Therefore, mitochondrial dysfunction caused by environmental damage, aging, and
especially DNA mutations may contribute to various human diseases3,4. While a cell’s DNA is located mainly in the nucleus, mitochondria contain their own DNA, known as mitochondrial DNA
(mtDNA)5. mtDNA plays an essential role in cellular respiration and energy consumption in humans and mice6. It is a 16.5-kb circular double-stranded DNA that encodes proteins for the
oxidative phosphorylation system, tRNAs, and ribosomal RNAs to aid in mitochondrial maintenance7,8. Unlike other organelles, mitochondria are enveloped by two distinct membranes, an outer
membrane, and an inner membrane, with an intermembrane area between them5. Additionally, there are cristae (folds in the inner membrane), which are crucial for ATP production9, and a matrix,
which is the space within the inner membrane. ATP produced in the mitochondria is synthesized by electron transferases in the inner membrane. These electron transferases include complex I
(NADH/ubiquinone oxidoreductase), complex II (succinate dehydrogenase), complex III (cytochrome c reductase), complex IV (cytochrome c oxidase), and complex V (ATP synthase) in the
mitochondrial cristae. Among them, mitochondrial complex I, which is composed of seven polypeptides (_MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND4L, MT-ND5_, and _MT-ND6_), is pivotal in the
electron transport chain, a process essential for maintaining the biological system10. Therefore, mutations in complex I genes may lead to various human diseases11. Mok et al. and Cho et al.
successfully developed C-to-T and A-to-G base editing in the mitochondrial genome12,13, making it possible to induce programmable base editing in double-stranded mtDNA to demonstrate
phenotypic changes in animals. Previous studies have recently established mice and rat models for mtDNA point mutations with distinct phenotypes, including hunched posture, brain anatomy
abnormalities, mitochondrial morphology changes, and motor ability impairments14,15. Furthermore, heteroplasmic knockout of the mitochondrial gene, with a mutation load for _MT-ND5_ gene
knockdown of up to 46%, has been achieved16. However, comprehensive phenotypic assessments of mitochondrial gene knockouts have rarely been reported. Thus, this study investigated apparent
phenotypes and genotypes as well as brain- and fat-related phenotypes in heteroplasmic mitochondrial gene-knockout mice throughout their generation. MATERIALS AND METHODS MRNA PREPARATION We
used DdCBE-encoding plasmids from a previous study17. These mRNA templates were prepared via polymerase chain reaction (PCR) via Q5 High-Fidelity DNA Polymerase (M0492S, NEB, Ipswich, MA,
USA) with the following primers: F: 5′-CATCAATGGGCGTGGATAG-3′ and R: 5′-GACACCTACTCAGACAATGC-3′. mRNAs were synthesized via an in vitro RNA transcription kit (mMESSAGE mMACHINE T7 Ultra kit,
Invitrogen, Carlsbad, CA, USA) and purified via a MEGAclear kit (Invitrogen, Carlsbad, CA, USA). All procedures were conducted according to the manufacturer’s instructions. ANIMALS
Hormone-injected wild-type (WT) C57BL/6 N female C57BL/6 N males were mated, and embryos were collected. Female mice from the ICR strain were used as surrogate mothers. Because mitochondria
follow maternal inheritance, female mutant mice were used for breeding, and phenotype analysis was executed with male mutant mice. All the mice were maintained in a specific pathogen-free
facility under a 12-h dark‒light cycle at a temperature of 20–26 °C and a humidity of 40–60%. MICROINJECTION OF MOUSE ZYGOTES In preparation for zygote microinjection, 4–6-week-old C57BL/6 N
female mice were superovulated by injecting pregnant mare serum gonadotropin ([5 IU, Prospec, Ness-Ziona, Israel]) and human chorionic gonadotropin hormone (5 IU, Sigma‒Aldrich, Burlington,
MA, USA) intraperitoneally at 48-h intervals. For microinjection, a mixture containing left and right DdCBE-encoding mRNAs (each at 300 ng/μL) was diluted in diethylpyrocarbonate-treated
injection buffer (0.25 mM ethylenediaminetetraacetic acid [EDTA], 10 mM Tris; pH 7.4) and injected into the cytoplasm of zygotes via a Nikon ECLIPSE Ti micromanipulator and a FemtoJet 4i
microinjector (Eppendorf, Hamburg, Germany). The embryos were then cultured in microdrops of KSOM + AA (Millipore, Burlington, MA, USA) at 37 °C for 2 days in a humidified atmosphere
containing 5% carbon dioxide (CO2). Two-cell-stage embryos were then implanted into the oviducts of 0.5-dpc pseudopregnant surrogate mothers. GENOTYPING AND TARGETED DEEP SEQUENCING All the
assay methods used for genotyping were identical. Only the samples (blastocysts, toes, and feces) used as DNA templates for PCR differed. Blastocyst-stage embryos and tissues from pups were
incubated in lysis buffer (25 mM sodium hydroxide [NaOH], 0.2 mM EDTA; pH 10) at 95 °C for 20 min, after which the pH was adjusted to 7.4 via the addition of
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, free acids without pH adjustment) at a final concentration of 50 mM. For targeted deep sequencing, nested first PCR and second PCR
were performed to create a high-throughput sequencing library, after which the final index sequences were merged via Q5 DNA polymerase. The primers used are listed in Supplementary Table 1.
The library was subjected to paired-end read sequencing via a MiniSeq platform (Illumina, San Diego, CA, USA). In all the cases, the sequencing results were joined into a single fastqjoin
file and analyzed via the CRISPR RGEN tool (http://www.rgenome.net/). OXYGEN CONSUMPTION RATES The oxygen consumption rates were measured via a Seahorse XFe24 Analyzer (Agilent, Santa Clara,
CA, USA) following the manufacturer’s protocol. One hundred microliters of suspended cells (106 cells/ml) were seeded into Seahorse XF24 V28 PS cell culture microplates (Agilent, Santa
Clara, CA, USA) 16 h before measurements were taken. An analysis was performed using Seahorse XF DMEM pH 7.4 supplemented with 25 mM glucose and 1 mM sodium pyruvate (Agilent, Santa Clara,
CA, USA). XF cell mito stress tests were then performed using 1.5 mM oligomycin, 0.5 mM FCCP, and 0.5 mM rotenone + antimycin A (Seahorse XF Cell Mito Stress Test Kit 6 XF Assays, Agilent,
Santa Clara, CA, USA). WESTERN BLOTTING & IMMUNOBLOTTING Proteins were isolated from the brains of WT and mutant mice via a DNeasy Blood & Tissue Kit (QIAGEN, Venlo, Netherlands).
The concentration of the cleared lysate was measured via a Bradford protein assay (Bio-Rad, Hercules, CA, USA). Prepared protein samples (total protein = 20 µg) were loaded onto
SDS‒polyacrylamide gel electrophoresis (PAGE) gels with 6% acrylamide depending on the target protein and transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA,
USA). These membranes were immunoblotted with the following primary antibodies: anti-mt-ND5 (PA5-36600, Thermo Fisher Scientific, Carlsbad, MA, USA), anti-NDUFB7 (NADH dehydrogenase
[ubiquinone] 1 beta subcomplex subunit 7, sc-365552, Santa Cruz Biotechnology, Dallas, TX, USA), and anti-β-actin (SC-47778, Santa Cruz Biotechnology, Dallas, TX, USA). The membranes were
subsequently incubated with the following secondary antibodies: anti-rabbit (7074, Cell Signal, Danvers, MA, USA) and anti-mouse (7076, Cell Signal, Danvers, MA, USA). Specific protein
complexes were visualized via the Clarity Western ECL Substrate (Bio-Rad, CA, USA). The signals were detected with a ChemiDoc XRS+ system (Bio-Rad, Hercules, CA, USA). Band intensities were
quantified by densitometry via ImageJ (NIH) and normalized to that of β-actin. MEASUREMENT OF ATP CONCENTRATION ATP concentrations in the brain and liver were determined with an ATP Assay
Kit (ab83355, Abcam, Cambridge, UK) according to the manufacturer’s protocol. Brain and liver tissues (10 mg) were used in the assay. Tissues were homogenized and lysed in ATP buffer. After
lysis, the samples were centrifuged at 13,000 × _g_ for 5 min at 4 °C to remove insoluble material. They were then loaded into a 96-well plate (SPL, Seoul, Korea) and diluted in ATP assay
buffer at the recommended proportions following the manufacturer’s protocol. The plate was incubated at room temperature for 30 min in the dark. The optical density (OD) was then measured at
570 nm with a microplate reader. TRANSMISSION ELECTRON MICROSCOPY (TEM) The tissues were dissected and fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 2 h at 4 °C,
followed by postfixation with 1% osmium tetroxide for 2 h on ice. These tissues were then embedded in Epon 812 after dehydration in ethanol and propylene oxide. Polymerizations were
performed using pure resin at 70 °C for 2 days. An ultramicrotome (UltraCut-UCT, Leica, Wetzlar, Germany) was used to obtain 70-nm ultrathin sections, which were collected on 100-mesh copper
grids and stained with 2% uranyl acetate and lead citrate. The accelerating voltage for TEM (Technai G2 Spirit TWIN, FEI, Hillsboro, OR, USA) was 120 kV. Electron micrographs were obtained
at a magnification of ×8000. HISTOLOGICAL ANALYSIS Mouse fat tissues were fixed in 10% formalin after perfusion with saline. Whole brains were soaked in 4% paraformaldehyde (PFA) solution
for 5 days at 4 °C with shaking. The samples were embedded in paraffin and sectioned at a thickness of 5 μm. The sections were then stained with H&E. To visualize mid-brain structures
and detect apoptotic markers, the sections were stained with antibodies against the neuronal marker NeuN (ab177487, Abcam, Cambridge, UK) and the apoptotic marker cleaved caspase-3 (9664S,
Cell signal, Danvers, MA, USA). To investigate apoptotic regions in the hippocampus of a mutant brain, a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was
conducted via a TUNEL assay kit (DeadEndTM Colorimetric TUNEL System, Promega, Madison, WI, USA) according to the manufacturer’s instructions. Briefly, the sections were permeabilized with
20 µg/ml proteinase K for 15 min and blocked with 30% hydrogen peroxide for 5 min at room temperature. Apoptotic signals were visualized via DAB, and hematoxylin was used as a
counterstaining agent. FEAR CONDITIONING Fear conditioning was used to evaluate aversive learning and memory. On Day 1, the mice were transported to a testing room and left undisturbed for
30 min before measurement. A tone cue at 80 dB was presented for 30 s, and a mild 0.8-mA foot shock was administered during the last 2 s of tone presentation. After an intertrial interval of
210 s, the procedure was repeated. On Day 2, the same light as that used on Day 1 was provided without tone cues. The mice were observed for the presence or absence of a freezing response.
The acquired data were analyzed on the basis of International Mouse Phenotyping Consortium (IMPC) standards (www.mousephenotype.org) by tracking software (FZ; O’Hara & Co, Ltd., Tokyo,
Japan). OPEN FIELD TEST The mice were placed in a 60 × 60-cm automated open field box that was homogeneously illuminated at 80 lx and allowed to explore freely for 20 min. Movement distance
and speed in the arena were detected by a machine at 5-min intervals for 20 min and recorded via computer software. The acquired data were analyzed according to IMPC standards
(www.mousephenotype.org) by tracking software (TimeOFCR4; O’Hara & Co, Ltd., Tokyo, Japan). MEASUREMENTS OF BODY WEIGHT, BODY COMPOSITION, AND BODY TEMPERATURE Body weight was measured
on electronic scales (Sartorius, Gottingen, Germany) after zeroing at the same time every week. Body composition was determined via a dual-energy X-ray absorptiometer (DEXA, Lunar Piximus
II, GE Medical Systems, Chicago, IL, USA). The mice were anesthetized with 1.2% avertin before measurement, and body temperature was measured via a rectal thermometer. BLOOD SERUM ANALYSIS
Glucose, total cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglyceride, phospholipid, and phosphorous levels were measured via a HITACHI Automatic Analyzer
7020 (HITACHI, Ibaraki, Japan) following the manufacturer’s instructions. Samples for testing fasting blood sugar were obtained at 15–18 h after food restriction via retro-orbital blood
collection. All blood samples were incubated at room temperature for 30 min and centrifuged at 12,000 rpm for 15 min to obtain serum samples. INDIRECT CALORIMETRY For indirect calorimetry
assessment, the mice were housed separately in metabolic chambers (CLAMS double feeder cages, Columbus instruments, Columbus, OH, USA) and had free access to food and water. The oxygen
consumption rates and CO2 production rates were measured for 24 hours. The metabolic data were analyzed simultaneously. COLD CHALLENGE To assess the body temperature of the cold-exposed
mice, the mice were exposed to 6 °C in a slow-temperature chamber (DHIN02-0034, DBL, Seoul, Korea). Body temperature was measured with a rectal thermometer (Testo 925, Testo, Kirchzarten,
Germany). WHOLE MITOCHONDRIAL GENOME SEQUENCING For whole mitochondrial genome sequencing, genomic DNA was extracted via a DNeasy Blood & Tissue Kit (Qiagen, Venlo, Netherlands). mtDNA
was amplified via PCR via PrimeSTAR GXL polymerase (Takara, Shiga, Japan). Amplified mtDNA was subjected to tagmentation via a DNA prep kit (Illumina, San Diego, CA, USA) following the
manufacturer’s protocol. The library was subjected to paired-end sequencing via the MiniSeq platform (Illumina, San Diego, CA, USA). SINGLE-CELL GENOTYPING IN ORGANS For single-cell
genotyping, the organs were ground with a 10-ml syringe plunger, and the cells were divided using 40-μm cell strainers (93040, SPL, Seoul, Korea). Divided cells were added to DMEM (11965084,
Gibco, Grand Island, NY, USA) and counted with a cell counter (AMQAX2000, Invitrogen, Carlsbad, CA, USA). After calculation, the cells were diluted and transferred to 96-well plates at 0.3
cells/well. Finally, individual cells were harvested, and genotyping was executed. STATISTICAL ANALYSIS The data are presented as the means \(\pm \,\)SEM for biologically independent
samples, and _P_ values were calculated via Student’s two-tailed _t_ test. The following _P_ values were used: *_p_ < 0.05, **_p_ < 0.01, and ***_p_ < 0.0001. The statistical
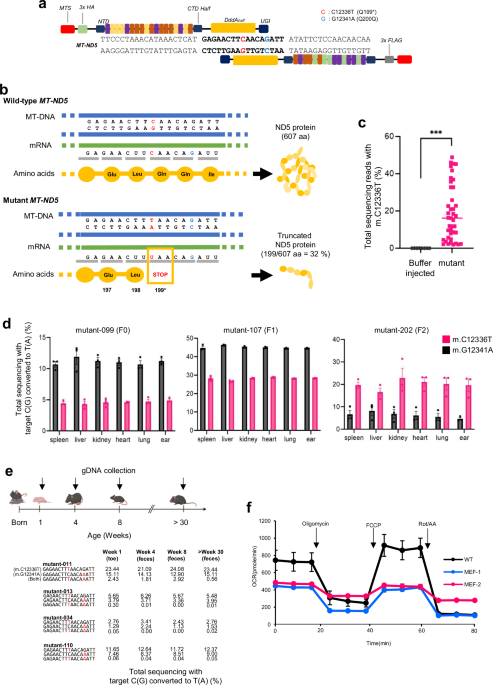
analysis was performed in Prism (GraphPad). RESULTS CONSTRUCTION OF _MT-ND5_ HETEROPLASMIC KNOCKOUT MUTANT MICE A nonsense mutation was created in the _MT-ND5_ gene of mice by microinjection
of DddA-derived cytosine base editor (DdCBE)-encoded mRNA into 1-cell-stage embryos. This induced the m.C12336T knockout mutation for incorporation of a premature stop codon and m.G12341A
bystander edit (silent mutation) in the editing window of _MT-ND5_ (Fig. 1a). We intended to achieve mtDNA editing for premature truncation at the 199th amino acid position in the mouse
_MT-ND5_ gene, where 5′-CAA-3′ glutamine was substituted with a 5′-TAA-3′ premature stop codon with a silent mutation in the 200th glutamine (Fig. 1b). Considering that mtDNA is maternally
inherited, we crossed F0 female mice with wild-type (WT) male mice to test germline transmission. We expanded the number of mice harboring the m.C12336T knockout mutation by breeding female
mutant mice. The results revealed that 2–48% of the generated mice harbored the m.C12336T knockout mutation (Fig. 1c). Moreover, these mutations were detected in various tissues of adult
mice (F0, mutant-099) at 14 weeks after birth, indicating the induction of a heteroplasmic knockout mtDNA mutation throughout the entire body of each mouse. These results were consistent
across the first, second, and third generations of mutant mice (Fig. 1d). These genes were confirmed by Sanger sequencing (Supplementary Fig. 1). Unexpectedly, the genotyping results of
individual cells in each organ revealed an extensive heteroplasmy range of the m.C12336T mutation. Furthermore, there was a cell with a 100% homoplasmy mutation rate in brain tissue, and the
median heteroplasmy rate of individual cells correlated with that of the whole-organ lysate (Supplementary Fig. 2). To verify the retention of editing frequency over time, we periodically
collected fecal samples from mutant mice and genotyped them via targeted deep sequencing. The frequencies of the m.C12336T and m.G12341A mutations remained stable for at least 30 weeks after
birth (Fig. 1e). The primary role of mitochondria is to generate energy by utilizing oxygen18. Thus, we assumed that mice with the induced knockout mutation may face challenges in ATP
production due to mitochondrial dysfunction. To validate the knockout mutation in the _MT-ND5_ gene in mice, we established a mouse embryonic fibroblast (MEF) cell line to assess the oxygen
consumption rates of the mutants. Compared with WT MEFs, mutant MEFs presented a significantly lower oxygen consumption rate (Fig. 1f), demonstrating impaired ATP synthesis due to
mitochondrial dysfunction in mutant mice. BRAIN DAMAGE IN _MT-ND5_ KNOCKOUT MICE Next, we aimed to investigate whether there was a loss-of-function mutation in the mtDNA necessary for the
function of specific organs, particularly those with high oxygen consumption, such as the brain. Despite constituting only ~2% of body weight, the brain consumes >20% of the body’s energy
needs in humans19. Organs with such high energy consumption are highly dependent on the proper functioning of mitochondria20. To determine the protein expression levels of MT-ND5 and
complex I in the brains of mutant mice, we conducted Western blot analyses for MT-ND5 and NDUFB7, which are known to be representative proteins identified via quantitative proteomic analysis
of complex I21. The protein levels of MT-ND5 and NDUFB7 were lower in all the mutant mice than in the WT mice (Fig. 2a). ATP levels in tissue lysates were also lower in mutant mice than in
WT mice (Fig. 2b). Additionally, TEM revealed damaged mitochondrial cristae in the cerebral cortex (Fig. 2c). To explore histopathological distinctions, we examined brain tissue sections
from both WT and mutant mice via hematoxylin and eosin (H&E) staining. There was no significant difference in the forebrain between WT and mutant mice (Supplementary Fig. 3). However,
mutant mice presented asymmetry in the hippocampus within midbrain regions (Fig. 2d). This asymmetry was observed more clearly via IHC with anti-NeuN (Supplementary Fig. 4). We observed a
1.6–2.4-fold difference in size between the left and right hippocampi of mutant mice at 14 weeks after birth (mutant-099 and mutant-104, respectively). Mice with hippocampal asymmetry showed
2.0–11.8% editing efficiency. Nevertheless, there was no statistically significant correlation between the mutant load and asymmetry ratio (Supplementary Fig. 5). Furthermore, intercell
spaces were wider in mutant mice than in WT mice, resulting in a thicker layer in the CA3 region of the abnormal hippocampus. Additionally, we observed that the number of apoptotic cells was
greater in the hippocampi of mutant mice than in those of WT mice via the TUNEL assay. We performed additional experiments with other apoptotic markers to detect apoptotic responses.
Compared with WT mice, mutant mice presented greater expression of cleaved caspase-3 (which is a general apoptotic marker) (Fig. 2d and Supplementary Fig. 4). Following these results, we
confirmed the occurrence of apoptosis in neuronal cells within the CA3 region of the hippocampus in mutant mice. The hippocampus plays a role in short-term memory, learning ability, and
behavioral activity by transmitting signals to other brain regions22,23. To investigate ethological changes in mutant mice, WT and mutant mice were subjected to an open-field test and fear
conditioning. The results showed that mutant mice tended to be hypoactive, demonstrating a 1.4-fold shorter total distance traveled and a 1.4-fold slower movement compared with WT mice.
There was also a 2.0-fold reduction in the center region dwell time, indicating that mutant mice had higher anxiety levels than WT mice. Additionally, we performed a fear conditioning test
for aversive learning by measuring freezing time by providing shocks and sound stimulation over 24-hour intervals. Only sound stimulation was provided in the second stimulation. Compared
with WT mice, heteroplasmic knockout mice presented a shorter freezing time, suggesting a fear-memorization disorder (Fig. 2e, f). Mice with abnormal behavior showed 2.0–21.1% editing
efficiency. However, a statistically significant correlation between the mutant load and ethological changes was not observed (Supplementary Fig. 6). FAT ACCUMULATION IN MUTANT MICE
Mitochondrial dysfunction resulting from mutations in mtDNA can lead to various metabolic disorders, such as maternally inherited diabetes and deafness (MIDD)24,25. The body weights of
mutant mice with an obese phenotype (mutant-Ob) were 1.2 times greater than those of WT mice (Fig. 3a, b), whereas not all mutant mice with the knockout genotype presented differences in
body weight. Five (29%) of the 17 mutant mice with an obesity-related phenotype presented an increase in body weight from 7 weeks after birth (Table 1). Encouraged by this result, we
measured the body compositions of mutant mice via dual-energy X-ray absorptiometry to assess the higher body weights observed in several mutant mice. Consequently, the average body weights
and fat masses of the mutant-Ob mice were 1.3- and 3.5-fold greater than those of the WT mice, respectively. The average body fat ratio (fat mass over body weight) of the mutant-Ob mice was
20% greater than that of the WT mice (Fig. 3c). We also conducted an autopsy of three different fat tissues from mutant-Ob mice to examine the abdominal, epididymal, and perirenal fat. These
tissues exhibited visible differences. Moreover, the weight of each fat tissue sample was significantly greater than that of the WT mouse tissue, with 3.7-, 4.4-, and 5.0-fold increases in
abdominal, epididymal, and perirenal fat, respectively (Fig. 3d). Furthermore, the sizes of the perirenal adipose cells in the mutant-Ob mice were measured via H&E-stained tissue
sections, which revealed that the degree of hypertrophy ranged between 1.9 and 4.4 times greater than that in the WT mice (Fig. 3e). Furthermore, TEM images revealed damaged mitochondrial
cristae in white adipose tissue (Fig. 3f). We also analyzed the levels of biochemical markers in the serum and ATP in white adipose tissue. While other serum markers were not significantly
different between the mutant-Ob and WT mice (Supplementary Fig. 7), the glucose levels in the mutant-Ob mice were twice those in the WT mice. Additionally, ATP levels were decreased in
mutant-Ob mice (Fig. 3g). Nevertheless, no difference was observed in the editing frequency of the m.C12336T mutation between mutant-Ob mice and those without obesity onset (Fig. 3h). We
assumed that _MT-ND5_ knockout mutant mice susceptible to obesity may be at risk of metabolic disorders. Thus, we examined the serum levels in WT, mutant-Ob, and mutant-non-Ob mice. In the
serum analysis, unlike in the mutant-Ob mice, blood glucose did not increase in the mutant-non-Ob mice, although a decrease in HDL was observed (Fig. 3i, j). HDL, considered instructive
cholesterol, has been reported to be low in diabetes patients26. However, there were no significant differences in the levels of cholesterol, LDL, triglycerides, phosphor lipids, or
phosphorous between the groups (Supplementary Fig. 7). These results suggest that the phenotypic expression and potential induction of obesity are attributed to heteroplasmic knockout
mutations. _MT-ND5_ NONSENSE MUTANTS ARE SUSCEPTIBLE TO OBESITY Encouraged by these results, we aimed to investigate differences between mutant-Ob and mutant mice not showing an obese
phenotype (mutant-non-Ob). Unlike mutant-Ob mice, mutant-non-Ob mice and WT mice presented no differences in external features, body weight (Fig. 4a, b), body composition (Fig. 4c), or
visible distinctions in fat tissue (Fig. 4d). However, several similarities were observed between the mutant-Ob and mutant-non-Ob mice. Similar to those in the mutant-Ob phenotypes, the
sizes of white adipose cells in the mutant-non-Ob mice were twice as large as those in the WT mice (Fig. 4e). Furthermore, damaged mitochondrial cristae were observed in white adipose
tissues from the mutant-non-Ob comparison (Fig. 4f). Additionally, ATP levels were similarly reduced in white adipose tissue lysates from mutant-non-Ob mice (Fig. 4g). To assess the
susceptibility to obesity, we fed a high-fat diet to mutant mice that did not exhibit the obese phenotype with MT-ND5 gene-knockdown mutations. Two weeks after the high-fat diet, the mutant
mice exhibited significantly greater weight gain than the WT mice (Fig. 4h). These results suggest that, regardless of the external features of obesity, mitochondrial heteroplasmic knockout
mice are highly susceptible to obesity. _MT-ND5_ NONSENSE MUTATION LEADS TO FAILURE OF HEAT PRODUCTION AND DECREASED THERMOREGULATION ABILITY One of the most essential functions of
mitochondria is thermoregulation, where brown adipose tissue (BAT), a mitochondria-rich tissue, modulates thermogenesis27. We hypothesized that mitochondrial heteroplasmic gene knockout
could induce thermogenic disorders. Thus, we conducted an indirect calorimetry test to assess the metabolic abilities of mutant mice. As shown in Fig. 5a, the oxygen consumption and CO2
production rates of the mutant mice were reduced during the dark cycle. Decreased metabolic ability led to the failure of heat production and abnormal drinking behavior (Fig. 5a). Assuming
that mutant mice have decreased heat production ability, we exposed both WT and mutant mice to a cold environment (6°C) to confirm abnormal thermogenesis in the mutant mice. We observed a
gradual decrease in rectal temperature over several hours. However, there were significant temperature decreases in the mutant mice after 5 hours. Furthermore, one mutant mouse died after 21
hours, and the remaining mutant mice also presented decreased rectal temperatures compared with those of WT mice (Fig. 5b). After the cold challenge, BATs from the mice were collected
through autopsy. As observed in white adipose tissues (Fig. 3), BATs of mutant mice appeared larger. These mice were 1.8 times heavier than the WT mice (Fig. 5c). The average cell size of
brown adipocytes from mutant mice was also 1.8 times larger than that of brown adipocytes from WT mice (Fig. 5d). Additionally, compared with WT mice, mutant mice presented decreased ATP
production in BAT (Fig. 5e). Damaged mitochondrial cristae were also observed in the BATs of the mutant mice (Fig. 5f). Taken together, these results demonstrated that mutant mice failed to
produce heat and exhibited impaired thermoregulation ability due to mitochondrial dysfunction induced by the _MT-ND5_ gene nonsense mutation. OFF-TARGET EFFECTS IN THE _MT-ND5_ NONSENSE
MUTANT A previous study revealed that DdCBE-mediated mtDNA editing could generate off-target activity in nuclear DNA14,28,29 and mitochondria30. Thus, we conducted whole-mitochondrial
genome-wide sequencing in mutant and mutant-Ob mice. The results revealed that mutant mice presented several off-target activities at the entire mtDNA locus with less than 5% editing
efficiency, and mutant-Ob mice presented only a few instances of off-target editing (Supplementary Fig. 8). Furthermore, we chose off-target sites harboring a perfect match or single
nucleotide mismatch with the left or right TALE binding sequence in the nuclear genome (Supplementary Table 2). Compared with WT mice, mutant and mutant-Ob mice, whose editing efficiency of
nonsense mutations ranged from 1% to 17%, presented negligible off-target editing frequencies (Supplementary Fig. 9). These results suggest that off-target editing has no apparent
relationship with phenotypic expression in mitochondrial knockout mutants. DISCUSSION In this study, we elucidated diverse phenotypes associated with the mitochondrial gene _MT-ND5_ knockout
mutation induced by the incorporation of a premature stop codon at the 199th position in the target gene. The heteroplasmic knockout, presumed to be a loss-of-function mutation, was
successfully achieved, with an editing frequency of up to 48%, and the desired mutations were observed in various tissues. Our findings demonstrate that microinjection of DdCBE-encoding mRNA
followed by consecutive crossings can yield sufficient _MT-ND5_ gene-knockdown mice for phenotype analysis without causing mtDNA copy number loss. Notably, these mice presented diverse
editing frequencies of the m.C12336T mutation, along with phenotypes related to metabolism and the brain. Mitochondrial dysfunction is recognized as the epicenter of energy metabolism. It
has been implicated in the pathophysiology of a variety of metabolic and neurodegenerative diseases that can affect vital organs, including the brain, heart, muscles, liver, eyes, and
pancreas31. In particular, mitochondria in the brain are essential for important biological processes, as they are involved in various biological functions, such as energy and free radical
production, cell metabolism, neurotransmitter regulation, apoptosis, and inflammation in the brain31,32,33. Recent studies have highlighted mitochondrial dysfunction as a significant
contributor to brain disorders, with hippocampal asymmetry being a notable feature in conditions such as Alzheimer’s disease and Parkinson’s disease34. We observed a simultaneous decrease in
the protein expression of MT-ND5 and complex I, as well as diminished ATP levels in the brains of mutant mice. Additionally, severe damage to the cristae of mitochondria was detected.
Mitochondria serve as primary sites for ATP production, where mitochondrial respiration chains, especially complex I, are involved in this process35. Furthermore, through histopathological
analysis, abnormalities in the hippocampus, including asymmetrical hippocampal atrophy and an increase in degenerated and apoptotic cells in CA3, were observed. Consequently, abnormalities
in learning memory were confirmed through behavioral assessment. Ethological changes can occur in mutant mice because the hippocampus plays a role in short-term memory, learning ability, and
behavioral activity by transmitting signals to other parts of the brain22,23. Specifically, intercell spaces were wider in mutant mice than in WT mice. The number of condensed nuclei and
apoptotic cells was greater in the hippocampi of mutant mice than in those of WT mice. Given the crucial role of the CA3 region in encoding spatial representations and episodic memories,
these results suggest that a nonsense mutation in the mitochondrial gene can lead to structural abnormalities in the brain with corresponding behavioral differences. This model could serve
as a valuable method for studying therapies for mitochondria-related brain diseases. Mitochondrial dysfunction may lead to the pathogenesis of metabolic disorders in tissues involved in
nutrient metabolism36. In particular, mitochondria in white adipose tissues are the main source of ATP and play critical roles in adipocyte biological processes such as adipogenesis,
lipolysis, and fatty acid oxidation37,38. We observed that 30% of the mutant mice exhibited a natural increase in body weight by 1.2-fold. These mice presented severely damaged mitochondria
in white adipose tissue, significantly reduced ATP levels, and substantial increases in body fat mass and blood glucose. Furthermore, in mutant-non-Ob mice, which do not naturally gain
weight, we observed severely damaged mitochondrial cristae, increased adipocyte size, and significantly reduced ATP and HDL levels. When these mutant-non-Ob mice were fed a high-fat diet,
they appeared to gain weight more rapidly than WT mice. These results demonstrate that MT-ND5 knockout can closely influence the metabolic obesity phenotype. Moreover, previous studies have
suggested that mitochondria in adipocytes play a role in regulating insulin sensitivity39. BrAT e specializes in consuming energy through heat production to maintain body temperature
homeostasis40. We observed damaged mitochondrial cristae in mutant BATs and a significant decrease in ATP levels. Moreover, compared with WT mice, mutant mice had difficulty maintaining body
temperature. Using our animal models, we demonstrated that mitochondria are essential organelles for maintaining the function of adipocytes in metabolic homeostasis, providing useful tools
for research on mitochondrial metabolic ability and disease. In this study, we conducted a comprehensive analysis of the phenotypes of heteroplasmic MT-ND5 gene-knockout mice, exploring the
therapeutic potential of MT-ND5 for human mitochondrial disorders. Initially, we hypothesized a correlation between the knockout genotype and the corresponding phenotype. However, in
knockouts with correction efficiencies ranging from 2% to 48%, there was no statistically significant difference between the knockdown mutation load and the extent of phenotypic expression,
unlike typical point mutations in the nuclear genome. We posit that mtDNA point mutation accumulation may surpass certain thresholds, eventually leading to phenotypic expression41,42,43.
Moreover, the well-known mitochondrial pathogenic mutation m.A3243G has been implicated not only in causing mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes
(MELAS) but also as a factor in MIDD44,45,46. According to recent reports, the reason that the same mutation can cause different disorders lies in heteroplasmy47,48. A lower heteroplasmy of
m.A3243G in MIDD patients than in MELAS patients has been reported47. Similarly, if the efficiency of m.C12336T exceeds 48%, there is also a possibility of another phenotype emerging.
Additionally, through single-cell analysis, we discovered that there is a variety of heteroplasmy at the cellular level, which suggests that even with the same editing efficiency,
differences in the distribution of single cells within the tissue could lead to variations in the degree of phenotypic expression. Several studies have reported that the average heteroplasmy
level of a single cell is determined by the proportion of mtDNA molecules with different heteroplasmy levels49,50. In our case, there was no statistical correlation between genotype and
phenotype expression in the obesity phenotypes (Figs. 3 and 4). However, the causes of the different phenotypic expression patterns in each individual are unclear. Based on several studies
on single-cell mtDNA49,50, we assumed that variations in the heteroplasmy level of single-cell mtDNA molecules could determine the expression of the phenotype. The phenotype could be
expressed when the heteroplasmy level of mtDNA molecules in a single cell is distributed largely at intermediate levels. However, when the deviation of mtDNA molecule heteroplasmy levels in
a single cell is large, the dysfunction of single cells harboring high levels of heteroplasmy mtDNA molecules could contribute to phenotypic expression. This phenomenon should be
investigated further and could be a great key for mitochondrial gene therapy. In conclusion, our findings contribute to the characterization of an animal model for heteroplasmic MT-ND5
gene-knockout mutation induced by programmable base editing in mtDNA. Our study revealed the harmful effects of mitochondrial gene knockout, which has not been previously reported (Fig. 6).
However, investigating specific and contextual mechanisms or pathways related to phenotype expression should be complemented by tissue- or cell-specific knockout systems. Taken together, our
results reveal the precise phenotypic features of mitochondrial heteroplasmic knockout, indicating that these mice could serve as valuable animal models for studying mitochondrial
dysfunction and developing therapies for human mitochondrial diseases. DATA AVAILABILITY Supplementary information is available in the online version of the paper. The data that support the
findings of this study are available from the corresponding author upon request. The high-throughput sequencing data from this study have been deposited in the NCBI Sequence Read Archive
(SRA) database under the accession codes PRJNA1172937. Source data are provided with this paper. REFERENCES * Willis, E. J. The powerhouse of the cell. _Ultrastruct. Pathol._ 16, iii–vi
(1992). Article PubMed CAS Google Scholar * Javadov, S., Kozlov, A. V. & Camara, A. K. S. Mitochondria in health and diseases. _Cells_ 9 (2020). https://doi.org/10.3390/cells9051177
* Wallace, D. C. Mitochondrial DNA in aging and disease. _Sci. Am._ 277, 40–47 (1997). Article PubMed CAS Google Scholar * DiMauro, S. & Schon, E. A. Mitochondrial DNA mutations in
human disease. _Am. J. Med. Genet._ 106, 18–26 (2001). Article PubMed CAS Google Scholar * Pittis, A. A. & Gabaldon, T. Late acquisition of mitochondria by a host with chimaeric
prokaryotic ancestry. _Nature_ 531, 101–104 (2016). Article PubMed PubMed Central CAS Google Scholar * Herzig, S. et al. Identification and functional expression of the mitochondrial
pyruvate carrier. _Science_ 337, 93–96 (2012). Article PubMed CAS Google Scholar * Anderson, S. et al. Sequence and organization of the human mitochondrial genome. _Nature_ 290, 457–465
(1981). Article PubMed CAS Google Scholar * Schon, K. R., Ratnaike, T., van den Ameele, J., Horvath, R. & Chinnery, P. F. Mitochondrial diseases: a diagnostic revolution. _Trends
Genet._ 36, 702–717 (2020). Article PubMed CAS Google Scholar * Giacomello, M., Pyakurel, A., Glytsou, C. & Scorrano, L. The cell biology of mitochondrial membrane dynamics. _Nat.
Rev. Mol. Cell Biol._ 21, 204–224 (2020). Article PubMed CAS Google Scholar * Sharma, L. K., Lu, J. & Bai, Y. Mitochondrial respiratory complex I: structure, function and implication
in human diseases. _Curr. Med. Chem._ 16, 1266–1277 (2009). Article PubMed PubMed Central CAS Google Scholar * MITOMAP: a human mitochondrial genome database. _Nucleic Acids Res._ 24,
177–179 (2019). * Mok, B. Y. et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. _Nature_ 583, 631–637 (2020). Article PubMed PubMed Central CAS
Google Scholar * Cho, S. I. et al. Targeted A-to-G base editing in human mitochondrial DNA with programmable deaminases. _Cell_ 185, 1764–1776.e1712 (2022). Article PubMed CAS Google
Scholar * Lee, S. et al. Enhanced mitochondrial DNA editing in mice using nuclear-exported TALE-linked deaminases and nucleases. _Genome Biol._ 23, 211 (2022). Article PubMed PubMed
Central CAS Google Scholar * Qi, X. et al. Precision modeling of mitochondrial disease in rats via DdCBE-mediated mtDNA editing. _Cell Discov._ 7, 95 (2021). Article PubMed PubMed
Central CAS Google Scholar * Lee, H. et al. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. _Nat. Commun._ 12, 1190 (2021). Article PubMed PubMed Central CAS
Google Scholar * Zaidi, A. A. et al. Bottleneck and selection in the germline and maternal age influence transmission of mitochondrial DNA in human pedigrees. _Proc. Natl. Acad. Sci. USA_
116, 25172–25178 (2019). Article PubMed PubMed Central CAS Google Scholar * Chan, D. C. Mitochondria: dynamic organelles in disease, aging, and development. _Cell_ 125, 1241–1252
(2006). Article PubMed CAS Google Scholar * Ambekar, T. et al. Mitochondrial quality control: epigenetic signatures and therapeutic strategies. _Neurochem. Int._ 148, 105095 (2021).
Article PubMed CAS Google Scholar * Shen, X., Sun, P., Zhang, H. & Yang, H. Mitochondrial quality control in the brain: the physiological and pathological roles. _Front. Neurosci._
16, 1075141 (2022). Article PubMed PubMed Central Google Scholar * Munday, D. C. et al. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus.
_Mol. Cell Proteom._ 9, 2438–2459 (2010). Article CAS Google Scholar * Eichenbaum, H. The hippocampus and declarative memory: cognitive mechanisms and neural codes. _Behav. Brain Res._
127, 199–207 (2001). Article PubMed CAS Google Scholar * Eichenbaum, H. Hippocampus: cognitive processes and neural representations that underlie declarative memory. _Neuron_ 44, 109–120
(2004). Article PubMed CAS Google Scholar * Murphy, R., Turnbull, D. M., Walker, M. & Hattersley, A. T. Clinical features, diagnosis and management of maternally inherited diabetes
and deafness (MIDD) associated with the 3243A>G mitochondrial point mutation. _Diabet. Med._ 25, 383–399 (2008). Article PubMed CAS Google Scholar * Malecki, M. T. et al. Maternally
inherited diabetes with deafness and obesity: body weight reduction response to treatment with insulin analogues. _Rev. Diabet. Stud._ 3, 205–207 (2006). Article PubMed Google Scholar *
Stadler, J. T. & Marsche, G. Obesity-related changes in high-density lipoprotein metabolism and function. _Int. J. Mol. Sci._ 21 (2020). https://doi.org/10.3390/ijms21238985 * Fenzl, A.
& Kiefer, F. W. Brown adipose tissue and thermogenesis. _Horm. Mol. Biol. Clin. Investig._ 19, 25–37 (2014). Article PubMed CAS Google Scholar * Lei, Z. et al. Mitochondrial base
editor induces substantial nuclear off-target mutations. _Nature_ 606, 804–811 (2022). Article PubMed CAS Google Scholar * Wei, Y. et al. Mitochondrial base editor DdCBE causes
substantial DNA off-target editing in nuclear genome of embryos. _Cell Discov._ 8, 27 (2022). Article PubMed PubMed Central CAS Google Scholar * Lee, S., Lee, H., Baek, G. & Kim, J.
S. Precision mitochondrial DNA editing with high-fidelity DddA-derived base editors. _Nat. Biotechnol._ 41, 378–386 (2023). Article PubMed CAS Google Scholar * Martin, L. J.
Mitochondrial and cell death mechanisms in neurodegenerative diseases. _Pharm. (Basel)_ 3, 839–915 (2010). CAS Google Scholar * Yin, F., Sancheti, H., Patil, I. & Cadenas, E. Energy
metabolism and inflammation in brain aging and Alzheimer’s disease. _Free Radic. Biol. Med_. 100, 108–122 (2016). Article PubMed PubMed Central CAS Google Scholar * Stefanatos, R. &
Sanz, A. The role of mitochondrial ROS in the aging brain. _FEBS Lett._ 592, 743–758 (2018). Article PubMed CAS Google Scholar * Claassen, D. O. et al. Cortical asymmetry in Parkinson’s
disease: early susceptibility of the left hemisphere. _Brain Behav._ 6, e00573 (2016). Article PubMed PubMed Central Google Scholar * Seo, J. H. et al. The mitochondrial
unfoldase-peptidase complex ClpXP controls bioenergetics stress and metastasis. _PLoS Biol._ 14, e1002507 (2016). Article PubMed PubMed Central Google Scholar * Bournat, J. C. &
Brown, C. W. Mitochondrial dysfunction in obesity. _Curr. Opin. Endocrinol. Diabetes Obes._ 17, 446–452 (2010). Article PubMed PubMed Central CAS Google Scholar * Gregoire, F. M., Smas,
C. M. & Sul, H. S. Understanding adipocyte differentiation. _Physiol. Rev._ 78, 783–809 (1998). Article PubMed CAS Google Scholar * Tormos, K. V. et al. Mitochondrial complex III
ROS regulate adipocyte differentiation. _Cell Metab._ 14, 537–544 (2011). Article PubMed PubMed Central CAS Google Scholar * Wang, C. H., Wang, C. C., Huang, H. C. & Wei, Y. H.
Mitochondrial dysfunction leads to impairment of insulin sensitivity and adiponectin secretion in adipocytes. _FEBS J._ 280, 1039–1050 (2013). Article PubMed CAS Google Scholar * Park,
A., Kim, W. K. & Bae, K. H. Distinction of white, beige and brown adipocytes derived from mesenchymal stem cells. _World J. Stem Cells_ 6, 33–42 (2014). Article PubMed PubMed Central
Google Scholar * Filograna, R., Mennuni, M., Alsina, D. & Larsson, N. G. Mitochondrial DNA copy number in human disease: the more the better? _FEBS Lett._ 595, 976–1002 (2021). Article
PubMed CAS Google Scholar * Chinnery, P. F., Samuels, D. C., Elson, J. & Turnbull, D. M. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is
there a common mechanism? _Lancet_ 360, 1323–1325 (2002). Article PubMed CAS Google Scholar * Rossignol, R. et al. Mitochondrial threshold effects. _Biochem. J._ 370, 751–762 (2003).
Article PubMed PubMed Central CAS Google Scholar * Li, R. & Guan, M. X. Human mitochondrial leucyl-tRNA synthetase corrects mitochondrial dysfunctions due to the tRNALeu(UUR) A3243G
mutation, associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like symptoms and diabetes. _Mol. Cell Biol._ 30, 2147–2154 (2010). Article PubMed PubMed Central
CAS Google Scholar * Suzuki, S. et al. Clinical features of diabetes mellitus with the mitochondrial DNA 3243 (A-G) mutation in Japanese: maternal inheritance and mitochondria-related
complications. _Diabetes Res. Clin. Pract._ 59, 207–217 (2003). Article PubMed CAS Google Scholar * Guillausseau, P. J. et al. Maternally inherited diabetes and deafness: a multicenter
study. _Ann. Intern. Med._ 134, 721–728 (2001). Article PubMed CAS Google Scholar * de Wit, H. M., Westeneng, H. J., van Engelen, B. G. & Mudde, A. H. MIDD or MELAS: that’s not the
question MIDD evolving into MELAS : a severe phenotype of the m.3243A>G mutation due to paternal co-inheritance of type 2 diabetes and a high heteroplasmy level. _Neth. J. Med._ 70,
460–462 (2012). PubMed Google Scholar * Wallace, D. C. & Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. _Cold Spring Harb. Perspect.
Biol._ 5, a021220 (2013). Article PubMed PubMed Central Google Scholar * Kotrys, A. V. et al. Single-cell analysis reveals context-dependent, cell-level selection of mtDNA. _Nature_ 629,
458–466 (2024). Article PubMed PubMed Central CAS Google Scholar * Floros, V. I. et al. Segregation of mitochondrial DNA heteroplasmy through a developmental genetic bottleneck in
human embryos. _Nat. Cell Biol._ 20, 144–151 (2018). Article PubMed PubMed Central CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the Korea Research
Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program (KGM1012311), grants (RS-2023-00261905 and RS-2023-00210965 and RS-2024-00408822 and RS-2024-00441068 and
RS-2024-00334994) of the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT), a Korea University Grant, and a Sungkyunkwan University (SKKU) Start-up Grant
(S-2023-1786-000-1). AUTHOR INFORMATION Author notes * These authors contributed equally: Sanghun Kim, Seul Gi Park. AUTHORS AND AFFILIATIONS * Department of Biomedical Sciences, Korea
University College of Medicine, Seoul, 02841, Republic of Korea Sanghun Kim, Jieun Kim, Sungjin Ju, Tae Young Jeong, Yeji Oh, Kyoungmi Kim & Hyunji Lee * Laboratory Animal Resource and
Research Center, Korea Research Institute of Bioscience and Biotechnology, Cheongju, 28116, Republic of Korea Seul Gi Park, Sang-Mi Cho, Eun-Kyoung Kim, Seunghun Han, Hae-Rim Kim, Taek Chang
Lee, Hyoung-Chin Kim, Won Kee Yoon, Ki-Hoan Nam & Hyunji Lee * Laboratory of Developmental Biology and Genomics, Research Institute for Veterinary Science, and BK21 PLUS Program for
Creative Veterinary Science Research, College of Veterinary Medicine, Seoul National University, Seoul, 08826, Republic of Korea Seongho Hong, Sol Pin Kim & Je Kyung Seong * Korea Model
animal Priority Center, Seoul National University, Seoul, 08826, Republic of Korea Seongho Hong, Soo-Yeon Lim, Su Bin Lee, Sol Pin Kim & Je Kyung Seong * Department of Physiology, Korea
University College of Medicine, Seoul, 02841, Republic of Korea Sungjin Ju, Tae Young Jeong & Kyoungmi Kim * Metabolic Regulation Research Center, Korea Research Institute of Bioscience
and Biotechnology (KRIBB), Daejeon, 34141, Republic of Korea Tae Hyeon An & Kyoung-jin Oh * Department of Functional Genomics, KRIBB School of Bioscience, Korea University of Science and
Technology (UST), Daejeon, 34141, Republic of Korea Tae Hyeon An & Kyoung-jin Oh * Department of MetaBioHealth, Sungkyunkwan University (SKKU), Suwon, 16419, Republic of Korea Seonghyun
Lee * Department of Precision Medicine, School of Medicine, Sungkyunkwan University (SKKU), Suwon, 16419, Republic of Korea Seonghyun Lee * Department of Convergence Medicine, Korea
University College of Medicine, Seoul, 02708, Republic of Korea Hyunji Lee Authors * Sanghun Kim View author publications You can also search for this author inPubMed Google Scholar * Seul
Gi Park View author publications You can also search for this author inPubMed Google Scholar * Jieun Kim View author publications You can also search for this author inPubMed Google Scholar
* Seongho Hong View author publications You can also search for this author inPubMed Google Scholar * Sang-Mi Cho View author publications You can also search for this author inPubMed Google
Scholar * Soo-Yeon Lim View author publications You can also search for this author inPubMed Google Scholar * Eun-Kyoung Kim View author publications You can also search for this author
inPubMed Google Scholar * Sungjin Ju View author publications You can also search for this author inPubMed Google Scholar * Su Bin Lee View author publications You can also search for this
author inPubMed Google Scholar * Sol Pin Kim View author publications You can also search for this author inPubMed Google Scholar * Tae Young Jeong View author publications You can also
search for this author inPubMed Google Scholar * Yeji Oh View author publications You can also search for this author inPubMed Google Scholar * Seunghun Han View author publications You can
also search for this author inPubMed Google Scholar * Hae-Rim Kim View author publications You can also search for this author inPubMed Google Scholar * Taek Chang Lee View author
publications You can also search for this author inPubMed Google Scholar * Hyoung-Chin Kim View author publications You can also search for this author inPubMed Google Scholar * Won Kee Yoon
View author publications You can also search for this author inPubMed Google Scholar * Tae Hyeon An View author publications You can also search for this author inPubMed Google Scholar *
Kyoung-jin Oh View author publications You can also search for this author inPubMed Google Scholar * Ki-Hoan Nam View author publications You can also search for this author inPubMed Google
Scholar * Seonghyun Lee View author publications You can also search for this author inPubMed Google Scholar * Kyoungmi Kim View author publications You can also search for this author
inPubMed Google Scholar * Je Kyung Seong View author publications You can also search for this author inPubMed Google Scholar * Hyunji Lee View author publications You can also search for
this author inPubMed Google Scholar CONTRIBUTIONS H.L. supervised the research. S.L. and H.L. conceived the research. S.K., S.G.P., S.L., K.K., J.K.S., and H.L. designed the study. S.K.,
S.G.P., S.L., K.K., J.K.S., and H.L. performed and analyzed the main experiments. J.K., S.H., S.-M.C., S.-Y.L., E.-K.K., S.J., S.B.L., S.P.K., S.H., H.-R.K., T.C.L., H.-C.K., W.K.Y., T.H.A.,
K.-J.O. and K.-H.N. performed the experiments. S.K., S.G.P., S.L., K.K., J.K.S., and H.L. wrote the manuscript. CORRESPONDING AUTHORS Correspondence to Ki-Hoan Nam, Seonghyun Lee, Kyoungmi
Kim, Je Kyung Seong or Hyunji Lee. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral
with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS OPEN ACCESS This article is
licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give
appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in
this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative
Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a
copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kim, S., Park, S.G., Kim, J. _et al._ Comprehensive
phenotypic assessment of nonsense mutations in mitochondrial ND5 in mice. _Exp Mol Med_ 56, 2395–2408 (2024). https://doi.org/10.1038/s12276-024-01333-9 Download citation * Received: 04
January 2024 * Revised: 07 July 2024 * Accepted: 30 July 2024 * Published: 01 November 2024 * Issue Date: November 2024 * DOI: https://doi.org/10.1038/s12276-024-01333-9 SHARE THIS ARTICLE
Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided
by the Springer Nature SharedIt content-sharing initiative
