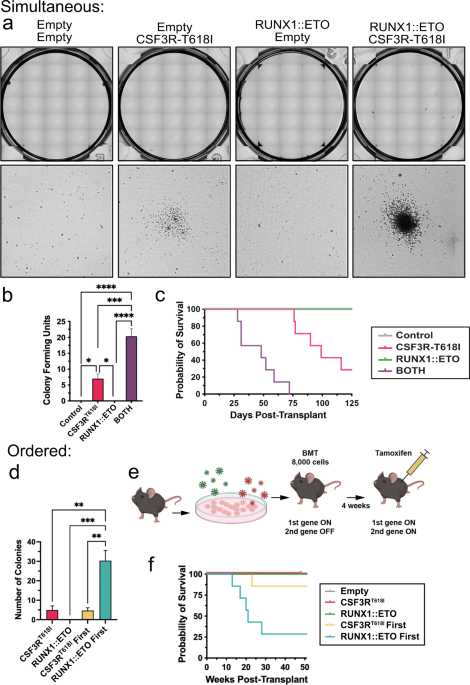
Runx1::eto translocations must precede csf3r mutations to promote acute myeloid leukemia development
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

Access through your institution Buy or subscribe Acute myeloid leukemia (AML) is a deadly blood cancer characterized by an overproduction of immature myeloid cells. AML has a high degree of
genetic diversity, and the presence or absence of particular genetic alterations can greatly impact prognosis. One such prognosis-altering mutation is in Colony Stimulating Factor 3 Receptor
(_CSF3R_, aka the G-CSF receptor). In pediatric AML, patients with _CSF3R_ mutations exhibit high relapse rates [1]. Similarly, in _CSF3R_-mutant adult AML, the 5-year overall survival rate
is a dismal 17% [2]. The role of _CSF3R_ in normal hematopoiesis is to induce a signaling cascade that results in the production of mature neutrophils. Constitutively activating mutations
in _CSF3R_ can drive chronic neutrophilic leukemia, a myeloproliferative neoplasm characterized by an abundance of mature neutrophils [3]. However, in the presence of a mutation that blocks
myeloid differentiation, _CSF3R_ mutations can also cause AML [4,5,6]. A recent study found that 90.5% of patients with _CSF3R_ mutations had co-occurring _RUNX1::ETO_ (8;21), _CBFB::MYH11_,
or _CEBPA_ mutations [4]. The _RUNX1::ETO_ and _CBFB::MYH11_ translocations disrupt the core binding factor complex, an important transcriptional regulator of myeloid development [7].
Similarly, mutations in the transcription factor _CEBPA_ impair differentiation to transform the myeloid lineage [8]. In prior studies, we found that mutations in _CEBPA_ block the ability
of _CSF3R_ to activate myeloid lineage enhancers without impairing _CSF3R_-driven proliferation programs [9]. This results in the expansion of myeloblasts that characterizes AML [9]. This is
a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per
year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support DATA AVAILABILITY Contact Julia Maxson at
[email protected] for information regarding renewable materials, datasets, and protocols. Genomic data are deposited at GEO with accession number GSE218829. REFERENCES * Tarlock K, Alonzo T,
Wang YC, Gerbing RB, Ries RE, Hylkema T, et al. Prognostic impact of CSF3R mutations in favorable risk childhood acute myeloid leukemia. Blood. 2020;135:1603–6. Article PubMed PubMed
Central Google Scholar * Su L, Gao S, Tan Y, Lin H, Liu X, Liu S, et al. CSF3R mutations were associated with an unfavorable prognosis in patients with acute myeloid leukemia with CEBPA
double mutations. Ann Hematol. 2019;98:1641–6. Article CAS PubMed Google Scholar * Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, et al. Oncogenic CSF3R mutations in
chronic neutrophilic leukemia and atypical CML. N Engl J Med. 2013;368:1781–90. Article CAS PubMed PubMed Central Google Scholar * Zhang Y, Wang F, Chen X, Zhang Y, Wang M, Liu H, et
al. CSF3R mutations are frequently associated with abnormalities of RUNX1, CBFB, CEBPA, and NPM1 genes in acute myeloid leukemia. Cancer. 2018;124:3329–38. Article CAS PubMed Google
Scholar * Sano H, Ohki K, Park MJ, Shiba N, Hara Y, Sotomatsu M, et al. CSF3R and CALR mutations in paediatric myeloid disorders and the association of CSF3R mutations with translocations,
including t(8; 21). Br J Haematol. 2015;170:391–7. Article CAS PubMed Google Scholar * Maxson JE, Ries RE, Wang YC, Gerbing RB, Kolb EA, Thompson SL, et al. CSF3R mutations have a high
degree of overlap with CEBPA mutations in pediatric AML. Blood. 2016;127:3094–8. Article PubMed PubMed Central Google Scholar * Speck NA, Gilliland DG. Core-binding factors in
haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–13. Article CAS PubMed Google Scholar * Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al.
Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–70. Article CAS PubMed Google Scholar
* Braun TP, Okhovat M, Coblentz C, Carratt SA, Foley A, Schonrock Z, et al. Myeloid lineage enhancers drive oncogene synergy in CEBPA/CSF3R mutant acute myeloid leukemia. Nat Commun.
2019;10:5455. Article PubMed PubMed Central Google Scholar * Wiemels JL, Xiao Z, Buffler PA, Maia AT, Ma X, Dicks BM, et al. In utero origin of t(8;21) AML1-ETO translocations in
childhood acute myeloid leukemia. Blood. 2002;99:3801–5. Article CAS PubMed Google Scholar * Kohzaki H, Ito K, Huang G, Wee HJ, Murakami Y, Ito Y. Block of granulocytic differentiation
of 32Dcl3 cells by AML1/ETO(MTG8) but not by highly expressed Bcl-2. Oncogene. 1999;18:4055–62. Article CAS PubMed Google Scholar * Metcalf D, Nicola NA. Proliferative effects of
purified granulocyte colony-stimulating factor (G-CSF) on normal mouse hemopoietic cells. J Cell Physiol. 1983;116:198–206. Article CAS PubMed Google Scholar * Wang GG, Calvo KR,
Pasillas MP, Sykes DB, Hacker H, Kamps MP. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–93. Article CAS PubMed Google
Scholar * Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. * Su L, Tan Y, Lin H,
Liu X, Yu L, Yang Y, et al. Mutational spectrum of acute myeloid leukemia patients with double CEBPA mutations based on next-generation sequencing and its prognostic significance.
Oncotarget. 2018;9:24970–9. Article PubMed PubMed Central Google Scholar * Guo H, Ma O, Speck NA, Friedman AD. Runx1 deletion or dominant inhibition reduces Cebpa transcription via
conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis. Blood. 2012;119:4408–18. Article CAS PubMed PubMed Central Google Scholar * Wang W, Wang X, Ward
AC, Touw IP, Friedman AD. C/EBPalpha and G-CSF receptor signals cooperate to induce the myeloperoxidase and neutrophil elastase genes. Leukemia. 2001;15:779–86. Article CAS PubMed Google
Scholar * Ma O, Hong S, Guo H, Ghiaur G, Friedman AD. Granulopoiesis requires increased C/EBPalpha compared to monopoiesis, correlated with elevated Cebpa in immature G-CSF receptor versus
M-CSF receptor expressing cells. PLoS One. 2014;9:e95784. Article PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS Research reported in this publication was
supported by NCI F32CA239422 and an OHSU SciOps award to SAC; an American Society of Hematology Research Restart Award, an American Society of Hematology Scholar Award and 1 K08 CA245224
from NCI awarded to TPB; NCI R01 CA247943, American Cancer Society RSG-19-184-01-LIB, and an LLS Scholar Award to JEM. We thank the following OHSU core facilities for their assistance:
Advanced Light Microscopy, Histopathology Shared Resource, Flow Cytometry Shared Resource, ExaCloud Cluster Computational Resource, and the Advanced Computing Center. AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Division of Oncologic Sciences, Knight Cancer Institute, Oregon Health & Science University, Portland, OR, 97239, USA Sarah A. Carratt, Garth L. Kong, Cody
Coblentz, Zachary Schonrock, Lauren Maloney, Ben Weeder, Will Yashar, Rowan Callahan, Hunter Blaylock, Colin Coleman, Dan Coleman, Theodore P. Braun & Julia E. Maxson * Division of
Hematology & Medical Oncology, Oregon Health & Science University, Portland, OR, 97239, USA Theodore P. Braun & Julia E. Maxson * Cell, Developmental and Cancer Biology, Oregon
Health & Science University, Portland, OR, 97239, USA Julia E. Maxson Authors * Sarah A. Carratt View author publications You can also search for this author inPubMed Google Scholar *
Garth L. Kong View author publications You can also search for this author inPubMed Google Scholar * Cody Coblentz View author publications You can also search for this author inPubMed
Google Scholar * Zachary Schonrock View author publications You can also search for this author inPubMed Google Scholar * Lauren Maloney View author publications You can also search for this
author inPubMed Google Scholar * Ben Weeder View author publications You can also search for this author inPubMed Google Scholar * Will Yashar View author publications You can also search
for this author inPubMed Google Scholar * Rowan Callahan View author publications You can also search for this author inPubMed Google Scholar * Hunter Blaylock View author publications You
can also search for this author inPubMed Google Scholar * Colin Coleman View author publications You can also search for this author inPubMed Google Scholar * Dan Coleman View author
publications You can also search for this author inPubMed Google Scholar * Theodore P. Braun View author publications You can also search for this author inPubMed Google Scholar * Julia E.
Maxson View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Concept and design: SAC, TPB, JEM. In vitro experiments: SAC, LM, HB, C. Coleman,
DC, TPB, JEM. In vivo experiments: SAC, C Coblentz, ZS, TPB. Computational resources and analysis: GLK, BW, WY, RC, TPB. Analysis and interpretation of data: all authors. Writing, review and
revision of the manuscript: all authors. CORRESPONDING AUTHORS Correspondence to Theodore P. Braun or Julia E. Maxson. ETHICS DECLARATIONS COMPETING INTERESTS JEM discloses a collaboration
with Ionis pharmaceuticals, research funding from Gilead Sciences, Kura Oncology and Blueprint Medicines. WY is a former employee of Abreos Biosciences, Inc. and was compensated in part with
common stock options. Pursuant to the merger and reorganization agreement between Abreos Biosciences, Inc. and Fimafeng, Inc., WY surrendered all of his common stock options in March 2021.
The other authors do not have conflicts of interest, financial or otherwise. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in
published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FILES RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds
exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely
governed by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Carratt, S.A., Kong, G.L., Coblentz, C. _et al._
_RUNX1::ETO_ translocations must precede _CSF3R_ mutations to promote acute myeloid leukemia development. _Leukemia_ 37, 1141–1146 (2023). https://doi.org/10.1038/s41375-023-01862-8 Download
citation * Received: 19 December 2022 * Revised: 20 February 2023 * Accepted: 24 February 2023 * Published: 09 March 2023 * Issue Date: May 2023 * DOI:
https://doi.org/10.1038/s41375-023-01862-8 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
