Suppression of pancreatic cancer liver metastasis by secretion-deficient itih5
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT BACKGROUND Previously, we identified ITIH5 as a suppressor of pancreatic ductal adenocarcinoma (PDAC) metastasis in experimental models. Expression of ITIH5 correlated with
decreased cell motility, invasion and metastasis without significant inhibition of primary tumour growth. Here, we tested whether secretion of ITIH5 is required to suppress liver metastasis
and sought to understand the role of ITIH5 in human PDAC. METHODS We expressed mutant ITIH5 with deletion of the N-terminal secretion sequence (ITIH5Δs) in highly metastatic human PDAC cell
lines. We used a human tissue microarray (TMA) to compare ITIH5 levels in uninvolved pancreas, primary and metastatic PDAC. RESULTS Secretion-deficient ITIH5Δs was sufficient to suppress
liver metastasis. Similar to secreted ITIH5, expression of ITIH5Δs was associated with rounded cell morphology, reduced cell motility and reduction of liver metastasis. Expression of ITIH5
is low in both human primary PDAC and matched metastases. CONCLUSIONS Metastasis suppression by ITIH5 may be mediated by an intracellular mechanism. In human PDAC, loss of ITIH5 may be an
early event and ITIH5-low PDAC cells in primary tumours may be selected for liver metastasis. Further defining the ITIH5-mediated pathway in PDAC could establish future therapeutic
exploitation of this biology and reduce morbidity and mortality associated with PDAC metastasis. SIMILAR CONTENT BEING VIEWED BY OTHERS SCHLAFEN 5 AS A NOVEL THERAPEUTIC TARGET IN PANCREATIC
DUCTAL ADENOCARCINOMA Article 12 April 2021 IL-8 IS A NOVEL PROMETASTATIC CHEMOKINE IN INTRAHEPATIC CHOLANGIOCARCINOMA THAT INDUCES CXCR2-PI3K/AKT SIGNALING UPON CD97 ACTIVATION Article
Open access 31 October 2023 CONCERTED CELL AND IN VIVO SCREEN FOR PANCREATIC DUCTAL ADENOCARCINOMA (PDA) CHEMOTHERAPEUTICS Article Open access 26 November 2020 BACKGROUND Pancreatic ductal
adenocarcinoma (PDAC) is a deadly cancer, due in part to its early and aggressive metastasis to distant organs. Unfortunately, over half of all patients diagnosed with PDAC present with
disseminated disease at diagnosis, and 5-year survival is approximately 3%.1 Due to hepatic portal circulation, the liver is the most common site of PDAC metastasis. In order to better
understand the factors regulating how PDAC spreads to the liver, we designed an in vivo siRNA screen for suppressors of PDAC metastasis and identified inter-α-trypsin-inhibitor-5 (ITIH5) as
a suppressor of PDAC metastasis; knockdown of ITIH5 was correlated with increased migration, invasion and liver metastasis.2 Much remains to be understood about the role of ITIH5 in both
normal physiology and tumour biology. The extracellular matrix protein ITIH5 has been shown to be a tumour suppressor in many cancer types3,4,5,6,7,8,9,10 where expression is lost likely due
to hypermethylation of the promoter.11,12 Compellingly, ITIH5 expression in breast cancer is also found to be associated with a rounded cell morphology13 and decreased metastatic
efficacy.14 However, its mechanism of metastasis suppression in PDAC remains unknown. Inter-α-trypsin inhibitors (IαI) are secreted proteins with protease inhibitory activity and are thought
to play roles in stabilisation of the extracellular matrices.15,16,17 As ITIH5 is also secreted, we hypothesised that ITIH5 secretion would be required for suppression of metastatic
outgrowth of PDAC cells in the liver. To test this hypothesis, we generated a construct with deletion of the secretion signal at the N terminus of ITIH5. The non-secreted mutant ITIH5
(ITIH5Δs) was expressed in highly metastatic human PDAC cells (with very low endogenous expression of ITIH5) to examine the effects of ITIH5Δs on PDAC morphology, migration and metastasis.
Interestingly, ITIH5Δs, which cannot be secreted (i.e. retained intracellularly), had similar effects as secreted ITIH5 on PDAC cell morphology, motility and suppression of liver metastasis.
These data open the possibility that there may be a yet-undescribed role for intracellular ITIH5 in suppression of PDAC liver metastasis. Additionally, we demonstrate reduced ITIH5 levels
in human primary PDAC and matched metastases, suggesting that loss of ITIH5 is an early event in PDAC cancer progression. METHODS CELL LINES AND CELL CULTURE Human PDAC cell lines (SUIT2
derivative S2-007, MiaPaCa-2 and PANC-1) were cultured in DMEM/F12 (Gibco, New York, USA, Cat #11330-032) with 5% foetal bovine serum (FBS) (Atlanta Biologicals, Atlanta, GA, USA, Cat
#S11195), 1% glutamine (Gibco, #25030-081), 0.25% non-essential amino acids (Gibco, #11140-050) and 1.5 and 1.0 μg/mL Puromycin (Gibco, #A11138-03) in a humidified incubator at 37 °C. All
cell lines were verified free of _Mycoplasma spp_. contamination by a PCR-based assay (ATCC, Manassas, VA, USA, #30-1012K). Cell line identification was authenticated using STR profiling.
PLASMIDS AND VIRAL TRANSDUCTION The pReceiver-Lv121 vector (GeneCopoeia, Cat #EX-E1333-Lv121, 10152 bp) was used for viral transduction to express full-length ITIH5 (“ITI” ORF 2829 bp). The
N-terminal 18AA secretion sequence of ITIH5 was removed from this construct to create the deleted secretion ITIH5 (ITIH5Δs, “Δs”) mutant. Deletions and restriction sites were inserted using
classical directional cloning techniques18 and verified by restriction digest and sequencing. Expression of full-length ITIH5 and ITIH5Δs was compared to expression of ITIH5 from control
cells expressing control vector (“Con”). Lentiviral transduction was used to introduce the construct into PDAC cells. DNA was amplified using XL10-Gold Ultracompetent Cells (Agilent, Santa
Clara, CA, USA, #200315). Plasmids were transfected into packaging cells using 2 µg of DNA, 6 µl of Vira Power Packing Plasmid DNA (ThermoFisher, #K4975-00), and 4 µl of jetPRIME
Transfection reagent (Polyplus, Berkeley, CA, USA, #114-15) in 900 µl of Polyplus Buffer (Polyplus, #712-60). This solution was added to packaging cell media for transduction during
overnight incubation. An additional 1 ml of media was added to each well of the 6-well plate at 24 h and 48 h after transfection, the viral supernatant was collected, precleared using
centrifugation (5’ at 1000 rpm) and filtered through 0.45 low protein-binding filters and applied to cells with 4 μg/ml hexadimethrine bromide (Sigma, St. Louis, MO) at 20–30% confluency.
Transductions occurred for 3 days, and then cells rested in culture media before selection in puromycin for 4 days. TRYPAN BLUE CELL PROLIFERATION ASSAYS To measure cell proliferation in
vitro, 1.0 × 104 human PDAC cells were plated in a 6-well cell culture dish (Corning, Cat #07-200-83) and grown in culture conditions listed above. At days 1, 3, 7, 10 and 14, cells were
washed with 1× PBS (Gibco, #10010-023) and detached using trypsin-EDTA (Gibco, #25200-056) until cells were free-floating. Then, an equal volume of media with foetal bovine serum (FBS,
Atlanta Biologicals, #S11195) was used to create a single-cell suspension. The concentration of cells was determined using a haemocytometer (Reichert, Depew, NY, USA, Cat #Z359629). Both
live and dead cells were counted as distinguished by exclusion or passive uptake of trypan blue solution (Sigma, Cat #T8154). Experiments were conducted in triplicate and standard errors
were calculated. CELL MIGRATION (SCRATCH) ASSAYS PDAC cell lines were cultured using the conditions described above. Cells were plated in 6-well plates (Corning, Corning, NY, USA, Cat
#07-200-83) to near confluency after 48 h of growth in a humidified incubator at 37 °C (~1–2 million cells). Plates were scratched using a 200-μl filtered tip (Avant, Valley Park, MO, USA,
Cat#ARS-200) with the plate lid as a guide. Two vertical and equally spaced scratches were made in each well. Plates were then rinsed twice with phosphate-buffered saline (PBS), and culture
media was replaced before phase-contrast imaging using a Nikon Eclipse TS100 Inverted Microscope, QImaging QIClick monochrome CCD camera and Metamorph® software. Cells were allowed to
migrate for 8 h (S2-007) or 72 h (MIAPaCa-2) before reimaging and assessing for closure. Surface area of the open space between cells was measured using ImageJ® software19 and the Wound
Healing Tool macro.20 For each timepoint, three images per scratch and two scratches per experimental group were recorded for each experiment. All experiments were repeated at least three
times. IMMUNOPRECIPITATION FROM CONDITIONED MEDIA Cells were plated and allowed to attach and grow for 48 h. When cells reached approximately 60% confluency, media was replaced, and cells
were allowed to grow for 24–48 h. Media was collected in microcentrifuge tubes and spun down at 12,000 rpm for 3 min at 4 °C to remove debris. The supernatant was transferred to a clean
microcentrifuge tube (1 ml/sample), and samples were precleared by adding 10 μl protein A/G beads (Santa Cruz, Cat# SC-2003) for 1 h at 4 °C. Samples were centrifuged at 12,000 rpm for 1 min
at 4 °C before transferring the supernatant to a new pre-chilled tube. Then, 1 μl of antibody was added to each sample and rotated for 1 h at 4 °C. For control samples, rabbit/mouse IgG
(Cell Signaling, Rabbit #2729 or Mouse #5415) was used as appropriate. In all, 20 μl protein A/G beads were added to the mixture, and the sample was incubated with rotation for 4 h overnight
at 4°C. Immunoprecipitated proteins were collected using centrifugation at 12,000 rpm for 1 min at 4 °C. The supernatant was aspirated, and beads were washed in 500 μl of ice-cold lysis
buffer three times with centrifugation for 1 min at 12,000 rpm and 4 °C. Samples were eluted from the pellet with 20 μl of loading buffer and heated at 95–100 °C for 5 min. Beads were
collected by centrifuging at 12,000 rpm for 1 min. The supernatant was transferred to a gel for electrophoresis. IMMUNOBLOTTING Cells cultured in vitro were lysed using 1× RIPA buffer
(Millipore, Burlington, MA, USA, #20-188) with 1% protease-phosphatase inhibitor (Thermo, #78411) on ice for 15 min before centrifuging (13,000 RPM) for 15 min. Protein concentration was
determined using a BCA assay (Thermo, #23227). Whole-cell lysates were denatured using NuPAGE LDS Sample Buffer (Invitrogen, Carlsbad, CA, USA, #NP0007) at 95 °C for at least 5 min before
separation using either a 12% or 4–20% gradient SDS-PAGE gel (Bio-Rad, Hercules, CA, USA, #5671093) and transferred to PVDF membranes (Bio-Rad, #170-4156), blocked for at least 1 h using 5%
non-fat milk in Tris Buffered Saline with Tween-20 (TBST). Antibodies were incubated overnight at 4 °C (ITIH5, ThermoFisher, #PA5-24445) or for 1 h at room temperature (GAPDH, Cell
Signaling, Beverly, MA, USA, #2118). A complete list of antibody catalogue numbers is available in Supplemental Table 1. EXTRACELLULAR VESICLE ISOLATION Extracellular vesicle (EV)
purification was performed as previously described21 with minor modifications. Briefly, cells were grown to 70% confluence at 48 h after plating in 10-cm dishes and washed with serum-free
medium. Cells were incubated in 7 ml/dish of serum-free medium or EV-free medium for 36–48 h before harvesting media. Cells were centrifuged in conical tubes in table-top centrifuge with
swing bucket rotor at 300 × _g_ for 10 min at 4 °C and then transferred to fresh tubes on ice. The supernatant was centrifuged at 2000 × _g_ for 10 min at 4 °C. The supernatant was
transferred to autoclaved 500-ml Nalgene bottles and centrifuged at 16,500 × _g_ for 30 min at 4 °C. The supernatant was transferred to thin polymer open-top tubes (17 ml, Fisher, #03-126)
and spun at 110,000 × _g_ (24,300 rpm) in an ultracentrifuge using a swing bucket (SureSpin630 6 × 16) rotor for 1.5 h at 4 °C. The supernatant was removed, and pellets were resuspended in 1
ml of ice-cold PBS before centrifuging at 110,000 × _g_ (24,300 rpm) using a swing bucket (SureSpin630 6 × 16) rotor for 1.5 h at 4 °C. The supernatant was removed, and the pellet was
resuspended in 20–40 μl of PBS before lysis, protein quantification and western blotting. Particle size was measured using a Nanosight® device (settings: frames per second: 24.99,
calibration: 189 nm/pixel, blur: auto, detection threshold: 6 multi, min track length: auto, min expected size: 100 nm, temperature: 22.70 °C and viscosity: 0.94 cP) and detected the
following particle size: mean: 155 nm, mode: 158 nm and SD: 52 nm. CELL FRACTIONATION Cytoplasmic–nuclear fractions were separated according to the manufacturer’s instructions using the
NE-PER™ Nuclear and Cytoplasmic Extraction kit (ThermoFisher, Waltham, MA, USA, #78835). Membrane–cytoplasmic fractions were separated according to the manufacturer’s instructions using the
Mem-PER™ Plus Membrane Protein Extraction kit (ThermoFisher, #89842). IMMUNOCYTOCHEMISTRY Cells were grown in standard tissue culture conditions and plated onto 8-well chamber slides
(Lab-Tek, Grand Rapids, MI, USA, #177402) using culture media. Cells were fixed using 4% paraformaldehyde (PFA) in PBS, blocked with 1% bovine serum albumin (BSA, Fisher, #BP9703-100) in PBS
and primary antibodies were applied at 1:100, while secondary antibodies were applied at 1:800 (Alexa 594, #R37117, ThermoFisher). Nuclei were counterstained with DAPI in mounting medium
(Vectashield, Burlingame, CA, USA, #H-1200). NUCLEAR LOCALISATION SEQUENCE IDENTIFICATION Putative nuclear localisation signals were identified using a well-established and highly utilised
sequence prediction algorithm available through NLS Mapper software (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi).22,23 Cut-off scores were set at 4.0 searching the entire
amino acid sequence in FASTA format. EXPERIMENTAL METASTASIS ASSAY—INTRASPLENIC INJECTIONS To measure growth of disseminated pancreatic cancer in liver, PDAC cells were injected
intrasplenically into athymic Foxn1nu (Envigo, Indianapolis, IN, USA) mice and allowed to circulate and seed the liver via the portal circulation for 2 min. Next, a splenectomy was performed
to remove intrasplenic tumour burden and allow for outgrowth of liver metastases. These procedures were performed as previously described.2,24 For S2-007 and MIAPaCa-2 cells, 5 × 105 or 1 ×
106 cells were injected into 100 µl of Hanks Balanced Salt Solution (HBSS, Gibco, #14175-103) and allowed to grow for 4 or 8 weeks, respectively, or until the mouse was moribund and
required euthanasia. All animal studies were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center (IACUC #2017-2409). IMMUNOHISTOCHEMISTRY
Both lungs, all lobes of the liver and PDAC metastases therein were collected during necropsy and fixed in formalin (Fisher, #SF-98-4) overnight before embedding in paraffin. Slides were cut
on a microtome (5–7 μm) and deparaffinised before immunohistochemistry (IHC). Antigen retrieval was performed using citrate buffer, pH 6.0 (Thermo, #AP-9003-500) in a steamer for 40 min
before cooling and washing with PBS. Peroxidase blocking was achieved using 3% hydrogen peroxide, 5% normal horse serum (Invitrogen, #26050070) and 1% normal goat serum (Invitrogen,
#16210064) for protein blocking, and primary antibodies were diluted 1:100 in protein block solution. Secondary antibodies were diluted 1:500 in protein block, and colorimetric reaction was
accomplished using diaminobenzidine (DAB, Vector Laboratories #SK-4105). Counterstaining was accomplished using Gill’s haematoxylin #3 (Fisher, #NC9964763), and slides were dried and
coverslipped using Permount (Fisher, #SP-15-100). IHC QUANTIFICATION IHC staining for ITIH5 was assessed semi-quantitatively. Aperio AT2 Scanning Microscope (Leica Biosystems, Wetzlar,
Germany) was used to scan IHC-stained slides, and Aperio ImageScope software (Leica Biosystems) was used to assess the staining intensity (0+ lowest intensity, 3+ highest intensity) and
location (cytoplasmic or nuclear). Areas of interest were identified by two independent observers and validated by a liver pathologist (MO). STATISTICAL ANALYSIS Statistical analysis and
graphing were done using R software25 and Microsoft Excel. Data were normally distributed with equal variance unless otherwise noted. Experimental group means were compared using one-way
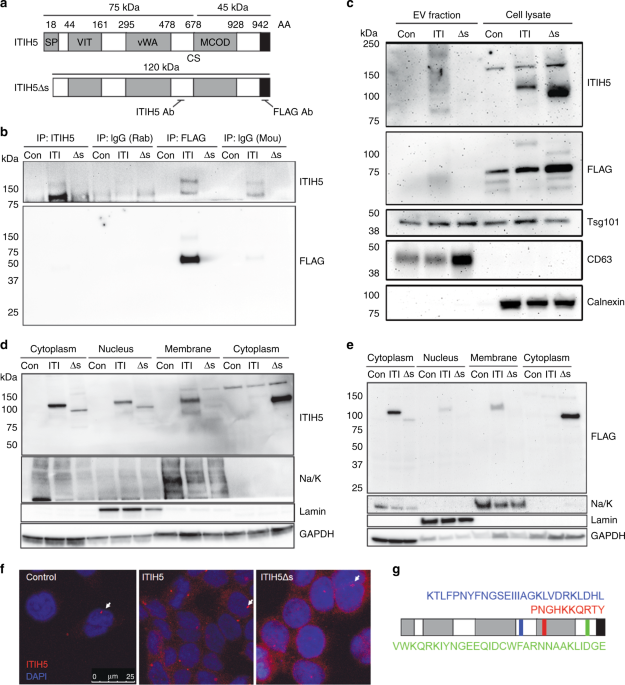
ANOVA. RESULTS EXPRESSION, LOCALISATION AND PEPTIDE PROCESSING OF ITIH5 IN HUMAN PDAC IαI family members may be post-translationally cleaved and covalently linked with chondroitin sulfate
proteoglycan (CSPG) or very short chains of hyaluronic acid (HA).15 Consistent with this pattern, ITIH5 contains a cleavage site (CS) at D681–V686,3 predicted to be cleaved by a number of
proteases.15 The ITIH5 antibody used in our experiments binds an epitope proximal to the CS, while FLAG epitope is at the C terminus of the protein (Fig. 1a). Thus, while both ITIH5 and FLAG
antibodies detect uncleaved ITIH5, ITIH5 and FLAG antibodies only detect the C-terminal or N-terminal fragments, respectively (Fig. 1b). Consistent with this reasoning, we detected
full-length and uncleaved ITIH5 (~120 kDa) with the ITIH5Δs construct at a slightly lower molecular weight due to the deletion of the 18AA secretion peptide. In the ITIH5 and ITIH5Δs groups,
we detected the C-terminal fragment at ~75 kDa (and ~150 kDa when two C-terminal fragments were presumably covalently linked by CSPG15), and an N-terminal fragment (~45 kDa) due to 17 AA
added after the ORF and a FLAG tag before the STOP codon in the expression construct (Fig. 1b). We expressed ITIH5 or ITIH5Δs constructs in three highly metastatic human PDAC cell lines that
express low endogenous ITIH5: S2-007, MiaPaCa-2 and Panc-1. Control cells express empty vector (“Con”), ITIH5-expressing cells express full-length ITIH5 (“ITI”) and ITIH5Δs cells express
secretion-deficient ITIH5 (“Δs”). Levels of ITIH5 overexpression were equivalent to endogenous ITIH5 measured in cultured pancreatic epithelial cells.2 As expected, non-secreted ITIH5Δs was
not found in conditioned media (Fig. 1b, S2-007 cells shown as representative), suggesting that removal of the secretion peptide prevented ITIH5 from being secreted extracellularly. Next, we
looked to see how ITIH5 might be secreted extracellularly by examining the contents of extracellular vesicles (EVs) as ITIH2 and ITIH3 can be secreted in EV.26 While both ITIH5 and ITIH5Δs
were expressed in the cell lysates (Fig. 1c), very little ITIH5 appeared to be secreted via EV (particle-size range 20–300 nm, mean 155 nm), suggesting that ITIH5 is secreted mainly by an
alternative pathway. To test our hypothesis that ITIH5 secretion would be required for suppression of pancreatic cancer metastasis, we first sought to characterise the intracellular
localisation of ITIH5 in PDAC. Using cell fractionation to separate cytoplasmic from nuclear or membrane fractions, we found that ITIH5 runs primarily at its full-length molecular weight of
120 kDa as shown using both the ITIH5 and FLAG antibodies (Fig. 1d, e). However, there are also detectable levels of ITIH5 that run at ~150 kDa, suggesting that ITIH5 is cleaved
intracellularly, and that two 75-kDa fragments may be linked by a short chain of CSPG or hyaluronic acid. When cytoplasmic and membrane fractions were separated, we observed consistently
less ITIH5Δs in the membrane fraction (Fig. 1d, e). While not formally tested here, this reduction in the amount of ITIH5Δs in the membrane fraction could be due to decreased trafficking
through the _trans_-Golgi causing reduced association with the membrane fraction, and/or decreased association with other membrane-associated proteins. Cytoplasmic and nuclear ITIH5 were
also observed by confocal immunofluorescence microscopy (IF) (Fig. 1f). Data suggesting nuclear localisation of ITIH5 were supported by the identification of three predicted nuclear
localisation signals for ITIH5, as predicted by amino acid sequence at 500–526AA, 731–741AA and 889–918AA (Fig. 1g). Which of these sequences might be responsible for nuclear localisation of
ITIH5/ITIH5Δs and the biological significance of nuclear ITIH5/ITIH5Δs is not yet understood and was not assessed in this report. EXPRESSION OF ITIH5 AFFECTS CELL MOTILITY AND MORPHOLOGY
Expression of ITIH5 can alter cell morphology in breast cancer.14 We observed similar changes in cell morphology in human PDAC after expressing either ITIH5 or non-secreted ITIH5Δs in three
metastatic human PDAC cell lines (S2-007, MIAPaCa-2 and Panc-1). All three of these cell lines share activating mutations in KRAS, inactivating mutations in p53 and inactivating mutations in
p16 (CDKN2A).27 Expression of ITIH5 was associated with a rounded cell morphology and dense aggregation of both S2-007 and MIAPaCa-2 human PDAC cells compared to control cells, with effects
in cells with the highest metastatic efficacy (S2-007) being most dramatic (Fig. 2a). Previously, we showed that expression of ITIH5 was correlated with decreased cell motility.2 We tested
the effect of expressing non-secreted ITIH5Δs on cell motility and found that intracellular ITIH5 also slowed coordinated cell motility in S2-007 (_P_ = 1.52e−7, Fig. 2b, c), although the
effects in MIAPaCa-2 were not statistically significant (Fig. 2d, e). The results measuring expression with respect to cell morphology and motility in Panc-1 were similar but less pronounced
(Supplemental Fig. 1a). We suspect that the reason we were unable to detect changes in cell migration rate in MIAPaCa-2 and Panc-1 is that these cell lines have an already slow rate of cell
migration in wound-healing assays. To confirm that this effect was truly a decrease in cell motility—and to reduce the likelihood that observed differences were due to ancillary processes
such as decreased rate of proliferation or increased rate of cell death—we measured proliferation in vitro and found that expression of either ITIH5 or ITIH5Δs did not significantly
proliferate cell death in any of the three human PDAC cell lines tested (S2-007, MIAPaCa-2 or Panc-1, Supplemental Fig. 1b). Next, we tested whether secretion of ITIH5 was necessary for
suppression of liver metastasis. We injected metastatic human PDAC S2-007 cells expressing ITIH5 or ITIH5Δs into 6- to 8-week-old nude mice intrasplenically and then allowed the cells to
seed the liver via the portal vein and grow in the liver for 4–8 weeks (_n_ = 18 mice per group). Intriguingly, non-secreted ITIH5Δs also suppressed liver metastasis (Fig. 3a). Animals
bearing disseminated tumours expressing ITIH5 or ITIH5Δs showed reduced number and size (measured by the longest dimension) of gross metastases as well as a reduced liver weight (Fig. 3b–d).
ITIH5- and ITIH5Δs-expressing cells developed approximately half the number of grossly visible metastases per liver compared to control mice (means: Con = 22.3, ITI = 11.4 and Δs = 9.17.
One-way ANOVA, _P_ = 0.0455). Similarly, ITIH5 and ITIH5Δs groups showed reduced average size of liver metastases (means: Con = 2.50 mm, ITI = 1.76 mm and Δs = 1.26 mm. One-way ANOVA, _P_ =
0.0012). In addition to showing a statistically significant difference in liver metastasis size, the ITIH5Δs group showed a statistically significant reduction in liver weight compared to
control (means: Con = 1.877 g, ITI = 1.643 g and Δs = 1.492 g. One-way ANOVA, _P_ = 0.0357). These data suggest an intracellular role for ITIH5 in mediating suppression of liver metastasis.
We next assessed livers and lungs from these mice (_n_ = 3 mice per group) histopathologically. All groups had microscopic liver metastases visible on histopathology (Fig. 3e), with ITIH5
and ITIH5Δs groups showing less liver area occupied by metastases histologically as compared to controls. Those results were not statistically significant (average ratio metastatic
area/total liver area, Con = 0.193, ITI = 0.059, Δs = 0.071. One-way ANOVA, _P_ > 0.05). Only control and ITIH5Δs groups had detected micrometastases in the lungs (Fig. 3f, average
metastases per image, Con = 1.21, Δs = 0.47, Student’s _t_ test, _P_ > 0.05). No gross metastases were visible on the surface of the lungs. Failure to detect lung metastases in the
ITIH5-injected mice does not exclude the possibility that full-length ITIH5 is more effective in suppressing lung metastasis than secretion-deficient ITIH5Δs. However, the most likely
interpretation is that the sample size was too small due to the low frequency of lung metastases formed by parental (control) PDAC cell lines. In addition to S2-007 cell lines, in vivo liver
metastasis experiments were also performed with MIAPaCa-2 cells, but injection of these cells failed to produce grossly visible liver and lung metastases in all groups (data not shown).
ITIH5 EXPRESSION IS LOW IN METASTASES IN VIVO AND IN HUMAN PDAC We evaluated protein levels of ITIH5 in liver metastases from mice following intrasplenic injection of S2-007 cells ±
expression of ITIH5 or ITIH5Δs by IHC. As anticipated, ITIH5 levels were higher in ITIH5- and ITIH5Δs-expressing tumours than in control tumours (Fig. 4a). ITIH5 or ITIH5Δs staining
intensities in liver metastases were high (2+ and 3+) compared to control cells (Fig. 4b). A small number of mice in the ITIH5 and ITIH5Δs groups developed large metastases. When expression
of ITIH5 was measured in small and large liver metastases, ITIH5 was high in small metastases (Fig. 4c) but low/lost in large metastases (Fig. 4c). To further understand clinical relevance,
we analysed ONCOMINE data sets and found that expression of ITIH5 mRNA in human PDAC was associated with lower grade, lower stage and with prolonged survival (Supplemental Fig. 1c, d). We
used a tissue microarray (TMA) and IHC to assess expression of ITIH5 in human PDAC samples (Fig. 4d). The array contained material from 38 patients, including matched primary and metastasis
(_n_ = 36 patients), two unmatched human primary and metastatic PDAC samples (_n_ = 1 each) and areas of uninvolved human pancreatic duct (_n_ = 11). Tissues used as normal controls came
from a total of 5 patients. Demographic data of patients included in the TMA are provided in Supplemental Table 2. The demographics of patients who died from PDAC and those who died differ,
likely to how their cause of death affected these demographic factors. Patients included as normal controls on the TMA died from the following causes: stroke (2), non-motor vehicle accident
(2) and motor vehicle accident (1). Survival duration data were unavailable for patients whose tissues were used as normal controls. In the TMA, IHC for ITIH5 revealed that ITIH5 levels in
metastases were similar to those in primary tumours (Fig. 4e). When ITIH5 levels in PDAC tumours were compared to normal pancreatic duct, staining intensity was lower in both primary and
metastatic tumours (Fig. 4e), as well as uninvolved normal pancreatic duct (Fig. 4f). These data may suggest that loss of ITIH5 expression may be an early event in PDAC progression, and
cells with reduced ITIH5 may be selected for liver metastasis. DISCUSSION ITIH5 is a secreted protein whose loss of expression has been described in many different cancers. In contrast to
our original hypothesis that secretion of ITIH5 is required for suppression of liver metastasis, we show that secretion of ITIH5 may not be necessary for suppression of liver metastasis in
PDAC. This exciting and unexpected result suggests that there may be a yet-undescribed mechanism for liver metastasis suppression by ITIH5. In contrast to other members of the
inter-α-trypsin family,28,29,30,31 ITIH5 does not appear to be secreted in EVs. While not formally tested here, like other proteins containing N-terminal secretion sequences, ITIH5 may be
passed through the endoplasmic reticulum (ER) via co-translational translocation and ultimately through the _trans_-Golgi through the constitutive secretory pathway.28,32,33 Removal of the
secretion peptide from ITIH5Δs could prevent translocation into the ER and account for the increased cytoplasmic retention observed with ITIH5Δs (Fig. 1d, e). We showed that expression of
both ITIH5 and secretion-deficient ITIH5Δs was correlated with an arrest of cell migration in PDAC. This biology may have a normal physiologic precedent as ITIH5 is highly expressed in the
placental syncytiotrophoblast,2 and in mice, this expression increases synchronously with arrest of invasion into the endometrium.34 Expression of ITIH5 has been associated with
transcriptional changes,13 and indeed, our data demonstrate that ITIH5 may be present in the nucleus, possibly implicating a role for ITIH5 in such changes. In the present study, we show a
reduction of ITIH5 expression in human PDAC using IHC. We did not, however, observe a further reduction in the levels of ITIH5 in metastatic lesions compared to primary tumours. This could
be a limitation of IHC quantification, or it may be that while ITIH5 transcription is reduced in more aggressive tumours, turnover of ITIH5 protein may be less dynamic and would thus fail to
show a similar reduction. Here, we provide what we believe to be the first IHC data for expression of ITIH5 in human PDAC. These results suggest that loss of ITIH5 expression is an early
event in PDAC progression, and cells with reduced ITIH5 may be selected for liver metastasis. The majority of PDAC cases show liver metastases at diagnosis,1 and many patients diagnosed with
localised disease eventually develop metastatic disease. Loss of ITIH5 in human PDAC may be one of many critical factors that induce PDAC metastases. In order to assess experimental liver
metastasis, we selected highly aggressive PDAC cell lines even though the intrasplenic model has limitations. For example, while we observed differences in the number and size of liver
metastases, we observed no statistically significant differences in the lung metastases between vector control cells and ITIH5-expressing cells. These results could be due to the lung being
more hospitable than liver,35 rendering growth-suppressive effects of ITIH5 less apparent. Alternatively, the metastasis-suppressing effects of ITIH5 could be liver-specific. In summary,
secretion of ITIH5 is not necessary for liver metastasis suppression in PDAC. There remains much to be understood about the biology of ITIH5 with respect to tumour biology. Understanding
this intracellular or cell-autonomous effect of ITIH5 and the pathways involved might help to uncover future therapeutic targets in order to treat patients with disseminated PDAC to the
liver and improve the outcome for most patients diagnosed with PDAC. Regardless of our incomplete knowledge of ITIH5 mechanism of action, the data reported here affirm the potential
relevance of ITIH5 as a biomarker for assessing tumour progression, metastasis and clinical outcomes for PDAC. REFERENCES * Siegel, R. L., Miller, K. D. & Jemal, A. Cancer Statistics,
2017. _CA Cancer J. Clin._ 67, 7–30 (2017). Article Google Scholar * Sasaki K., Kurahara H., Young E. D., Natsugoe S., Ijichi A., Iwakuma T. et al. Genome-wide in vivo RNAi screen
identifies ITIH5 as a metastasis suppressor in pancreatic cancer. _Clin. Exp. Metastasis_ 34, 229–239 (2017). * Himmelfarb, M., Klopocki, E., Grube, S., Staub, E., Klaman, I., Hinzmann, B.
et al. ITIH5, a novel member of the inter-alpha-trypsin inhibitor heavy chain family is downregulated in breast cancer. _Cancer Lett._ 204, 69–77 (2004). Article CAS Google Scholar *
Hamm, A., Veeck, J., Bektas, N., Wild, P. J. & Hartmann, A. Frequent expression loss of Inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a
systematic expression analysis. _BMC Cancer_ 8, 25 (2008). Article Google Scholar * Pita, J. M., Banito, A., Cavaco, B. M. & Leite, V. Gene expression profiling associated with the
progression to poorly differentiated thyroid carcinomas. _Br. J. Cancer_ 101, 1782–1791 (2009). Article CAS Google Scholar * Lu, Y., Liu, P., Wen, W., Grubbs, C. J., Townsend, R. R.,
Malone, J. P. et al. Cross-species comparison of orthologous gene expression in human bladder cancer and carcinogen-induced rodent models. _Am. J. Transl. Res._ 3, 8–27 (2010). PubMed
PubMed Central Google Scholar * Oing, C., Jost, E., Dahl, E., Wilop, S., Brummendorf, T. H. & Galm, O. Aberrant DNA hypermethylation of the ITIH5 tumor suppressor gene in acute myeloid
leukemia. _Clin. Epigenetics_ 2, 419–423 (2011). Article CAS Google Scholar * Zhang, S., Feng, X. L., Shi, L., Gong, C. J., He, Z. J., Wu, H. J. et al. Genome-wide analysis of DNA
methylation in tongue squamous cell carcinoma. _Oncol. Rep._ 29, 1819–1826 (2013). Article CAS Google Scholar * Kloten, V., Rose, M., Kaspar, S., von Stillfried, S., Knuchel, R. &
Dahl, E. Epigenetic inactivation of the novel candidate tumor suppressor gene ITIH5 in colon cancer predicts unfavorable overall survival in the CpG island methylator phenotype.
_Epigenetics_ 9, 1290–1301 (2014). Article Google Scholar * Perez-Ramirez, M., Hernandez-Jimenez, A. J., Guerrero-Guerrero, A., Benadon-Darszon, E., Perezpena-Diazconti, M., Siordia-Reyes,
A. G. et al. Genomics and epigenetics: A study of ependymomas in pediatric patients. _Clin. Neurol. Neurosurg._ 144, 53–58 (2016). Article Google Scholar * Veeck, J., Chorovicer, M.,
Naami, A., Breuer, E., Zafrakas, M., Bektas, N. et al. The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node-negative breast cancer and its aberrant expression
is caused by promoter hypermethylation. _Oncogene_ 27, 865–876 (2008). Article CAS Google Scholar * Rose, M., Bringezu, S., Godfrey, L., Fiedler, D., Gaisa, N. T., Koch, M. et al. ITIH5
and ECRG4 DNA methylation biomarker test (EI-BLA) for urine-based non-invasive detection of bladder cancer. _Int. J. Mol. Sci._ 21, 1117 (2020). Article CAS Google Scholar * Rose, M.,
Meurer, S. K., Kloten, V., Weiskirchen, R., Denecke, B., Antonopoulos, W. et al. ITIH5 induces a shift in TGF-beta superfamily signaling involving Endoglin and reduces risk for breast cancer
metastasis and tumor death. _Mol. Carcinog._ 57, 167–181 (2018). Article CAS Google Scholar * Rose, M., Kloten, V., Noetzel, E., Gola, L., Ehling, J., Heide, T. et al. ITIH5 mediates
epigenetic reprogramming of breast cancer cells. _Mol. Cancer_ 16, 44 (2017). Article Google Scholar * Zhuo, L. & Kimata, K. Structure and function of inter-alpha-trypsin inhibitor
heavy chains. _Connect Tissue Res_. 49, 311–320 (2008). Article CAS Google Scholar * Huang, L., Yoneda, M. & Kimata, K. A serum-derived hyaluronan-associated protein (SHAP) is the
heavy chain of the inter alpha-trypsin inhibitor. _J. Biol. Chem._ 268, 26725–26730 (1993). CAS PubMed Google Scholar * Zhao, M., Yoneda, M., Ohashi, Y., Kurono, S., Iwata, H., Ohnuki, Y.
et al. Evidence for the covalent binding of SHAP, heavy chains of inter-alpha-trypsin inhibitor, to hyaluronan. _J. Biol. Chem._ 270, 26657–26663 (1995). Article CAS Google Scholar *
Green M. R., Sambrook J. & Sambrook J. _Molecular Cloning: a Laboratory Manual_, 4th ed. (Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2012). * Schneider, C. A., Rasband,
W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. _Nat. Methods_ 9, 671–675 (2012). Article CAS Google Scholar * Baecker V. ImageJ macro tool sets for
biological image analysis. http://dev.mri.cnrs.fr/projects/imagej-macros/wiki/Wound_Healing_Tool. Accessed 12/12/2016, 2016 (2012). * Thery, C., Amigorena, S., Raposo, G. & Clayton, A.
Isolation and characterization of exosomes from cell culture supernatants and biological fluids. _Curr. Protoc. Cell Biol._ CHAPTER 3, Unit 3 22 (2006). PubMed Google Scholar * Kosugi, S.,
Hasebe, M., Entani, T., Takayama, S., Tomita, M. & Yanagawa, H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. _Chem.
Biol._ 15, 940–949 (2008). Article CAS Google Scholar * Kosugi, S., Hasebe, M., Tomita, M. & Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic
shuttling proteins by prediction of composite motifs. _Proc. Natl Acad. Sci. USA_ 106, 10171–10176 (2009). Article CAS Google Scholar * Kurahara, H., Bohl, C., Natsugoe, S., Nishizono,
Y., Harihar, S., Sharma, R. et al. Suppression of pancreatic cancer growth and metastasis by HMP19 identified through genome-wide shRNA screen. _Int. J. Cancer_ 139, 628–638 (2016). Article
CAS Google Scholar * R: A language and environment for statistical computing. (2015). * Linscheid C. R. _Expression, Regulation and Release of Human Placental Antigens: Implications for
the Maternal Immune Response to the Fetus_ [Dissertation]: (Anatomy and Cell Biology, University of Kansas, 2015) * Barretina, J., Caponigro, G., Stransky, N., Venkatesan, K., Margolin, A.
A., Kim, S. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. _Nature_ 483, 603–607 (2012). Article CAS Google Scholar * Sanders, S.
L., Whitfield, K. M., Vogel, J. P., Rose, M. D. & Schekman, R. W. Sec61p and BiP directly facilitate polypeptide translocation into the ER. _Cell_ 69, 353–365 (1992). Article CAS
Google Scholar * Swiatly, A., Horala, A., Hajduk, J., Matysiak, J., Nowak-Markwitz, E. & Kokot, Z. J. MALDI-TOF-MS analysis in discovery and identification of serum proteomic patterns
of ovarian cancer. _BMC Cancer_ 17, 472 (2017). Article Google Scholar * Li, X., Li, B., Li, B., Guo, T., Sun, Z., Li, X. et al. ITIH4: effective serum marker, early warning and diagnosis,
hepatocellular carcinoma. _Pathol. Oncol. Res_. 24, 663–670 (2018). Article CAS Google Scholar * Bost, F., Diarra-Mehrpour, M. & Martin, J. P. Inter-alpha-trypsin inhibitor
proteoglycan family-a group of proteins binding and stabilizing the extracellular matrix. _Eur. J. Biochem_. 252, 339–346 (1998). Article CAS Google Scholar * Rapoport, T. A. Protein
transport across the endoplasmic reticulum membrane: facts, models, mysteries. _FASEB J._ 5, 2792–2798 (1991). Article CAS Google Scholar * Kanner, E. M., Friedlander, M. & Simon, S.
M. Co-translational targeting and translocation of the amino terminus of opsin across the endoplasmic membrane requires GTP but not ATP. _J. Biol. Chem._ 278, 7920–7926 (2003). Article CAS
Google Scholar * Morcel, K., Watrin, T., Jaffre, F., Deschamps, S., Omilli, F., Pellerin, I. et al. Involvement of ITIH5, a candidate gene for congenital uterovaginal aplasia
(Mayer-Rokitansky-Kuster-Hauser syndrome), in female genital tract development. _Gene Expr._ 15, 207–214 (2012). Article Google Scholar * Zhong, Y., Macgregor-Das, A., Saunders, T.,
Whittle, M. C., Makohon-Moore, A., Kohutek, Z. A. et al. Mutant p53 together with TGFbeta signaling influence organ-specific hematogenous colonization patterns of pancreatic cancer. _Clin.
Cancer Res._ 23, 1607–1620 (2017). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank Hiroshi Kurahara, M.D. for initial experiments and scientific feedback,
Alejandro Parrales, Ph.D., Atul Ranjan, Ph.D., Swathi Iyer, Ph.D. and Adam Scheid, Ph.D. for scientific discussions and feedback on experiments. We acknowledge Ms. Elena Shergina and Ms.
Katie McKenzie for their discussions regarding statistical analysis. We also thank Dr. Lane Christensen for allowing us to use the Nanosight device and guidance during the EV experiments. We
would also like to acknowledge the University of Kansas Cancer Center Biospecimen Repository Core Facility for their effort in sample procurement. The authors would like to acknowledge Ms.
Valerie Freeman and Ms. Janet Fletcher for their administrative assistance. We thank Ms. Anca Geana and the University of Kansas Research Institute for assistance with grant management.
AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Cancer Biology, University of Kansas Medical Center, Kansas City, KS, USA Eric D. Young, Sharon J. Manley, Thomas C. Beadnell,
Alexander E. Shearin, Rosalyn Zimmerman, Evan Kauffman, Carolyn J. Vivian, Aishwarya Parasuram, Tomoo Iwakuma, Maura O’Neil & Danny R. Welch * Department of Digestive Surgery, Breast and
Thyroid Surgery, Kagoshima University, Kagoshima, Japan Ken Sasaki * Eppley Institute for Cancer Research, University of Nebraska Medical Center, Omaha, NE, USA Paul M. Grandgenett &
Michael A. Hollingsworth * Department of Pathology and Laboratory Medicine, University of Kansas Medical Center, Kansas City, KS, USA Danny R. Welch Authors * Eric D. Young View author
publications You can also search for this author inPubMed Google Scholar * Sharon J. Manley View author publications You can also search for this author inPubMed Google Scholar * Thomas C.
Beadnell View author publications You can also search for this author inPubMed Google Scholar * Alexander E. Shearin View author publications You can also search for this author inPubMed
Google Scholar * Ken Sasaki View author publications You can also search for this author inPubMed Google Scholar * Rosalyn Zimmerman View author publications You can also search for this
author inPubMed Google Scholar * Evan Kauffman View author publications You can also search for this author inPubMed Google Scholar * Carolyn J. Vivian View author publications You can also
search for this author inPubMed Google Scholar * Aishwarya Parasuram View author publications You can also search for this author inPubMed Google Scholar * Tomoo Iwakuma View author
publications You can also search for this author inPubMed Google Scholar * Paul M. Grandgenett View author publications You can also search for this author inPubMed Google Scholar * Michael
A. Hollingsworth View author publications You can also search for this author inPubMed Google Scholar * Maura O’Neil View author publications You can also search for this author inPubMed
Google Scholar * Danny R. Welch View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS E.D.Y. designed and planned experiments, performed data
analysis and drafted the paper. S.J.M. contributed to experimental design, performed and assisted with key experiments and revised the paper. T.C.B. performed data analysis and revised the
paper. A.E.S. developed experimental protocols, performed key experiments and revised the paper. K.S. developed experimental protocols and reagents, designed and assisted with experiments
and edited the paper. R.Z. designed and performed key experiments and revised the paper. E.K. performed key experiments. C.J.V. developed experimental protocols and reagents and revised the
paper. A.P. performed key experiments. T.I. assisted in designing experiments, troubleshooting experiments and revised the paper. P.M.G. developed reagents and reviewed the paper. M.A.H.
developed reagents and reviewed the paper. M.O. helped design and perform experiments, perform data analysis and reviewed the paper. D.R.W. designed experiments, drafted and revised the
paper. CORRESPONDING AUTHOR Correspondence to Danny R. Welch. ETHICS DECLARATIONS ETHICS APPROVAL AND CONSENT TO PARTICIPATE All animal studies were approved by the Institutional Animal Care
and Use Committee at the University of Kansas Medical Center (IACUC #2017-2409). Any studies using tissue solely obtained from the Rapid Autopsy Program for Pancreas (IRB#091-01) are not
considered human subject research because the individuals are not living when the tissue is obtained (per the regulations at 45 CFR 46.102) and therefore would not need IRB approval. CONSENT
TO PUBLISH Not applicable. DATA AVAILABILITY Data supporting the results can be found in Supplemental Fig. 1. A comprehensive list of antibodies used in this study can be found in
Supplemental Table 1. COMPETING INTERESTS The authors declare no competing interests. FUNDING INFORMATION The authors of this paper have research support from the NIH F30CA216998 (E.D.Y.),
Kansas INBRE, P20 GM103418 (S.J.M. and D.R.W.), DOD BCRP BC171381 (T.C.B.), the University of Kansas Cancer Center Summer Student Research Training Program (A.E.S.), the Natl. Fndn. Cancer
Res., CA168524, Hall Family Professorship in Molecular Medicine (D.R.W.), SPORE in Pancreatic Cancer, CA127297, TMEN Tumor Microenvironment Network, U54CA163120 and CA36727 (P.M.G. and
M.A.H.). ADDITIONAL INFORMATION NOTE This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will
switch to a Creative Commons Attribution 4.0 International (CC BY 4.0). PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and
institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTAL MATERIALS (COMPOSITE) RIGHTS AND PERMISSIONS This article is licensed under a Creative Commons Attribution 4.0
International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the
source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's
Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not
permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Young, E.D., Manley, S.J., Beadnell, T.C. _et al._ Suppression of pancreatic cancer
liver metastasis by secretion-deficient ITIH5. _Br J Cancer_ 124, 166–175 (2021). https://doi.org/10.1038/s41416-020-01093-z Download citation * Received: 04 March 2020 * Revised: 14 August
2020 * Accepted: 03 September 2020 * Published: 07 October 2020 * Issue Date: 05 January 2021 * DOI: https://doi.org/10.1038/s41416-020-01093-z SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative
