Measurable residual disease analysis in paediatric acute lymphoblastic leukaemia patients with abl-class fusions
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT BACKGROUND ABL-class fusions including _NUP214-ABL1_ and _EBF1-PDGFRB_ occur in high risk acute lymphoblastic leukaemia (ALL) with gene expression patterns similar to
_BCR-ABL_-positive ALL. Our aim was to evaluate new DNA-based measurable residual disease (MRD) tests detecting these fusions and _IKZF1_-deletions in comparison with conventional
immunoglobulin/T-cell receptor (Ig/TCR) markers. METHODS Precise genomic breakpoints were defined from targeted or whole genome next generation sequencing for ABL-fusions and _BCR-ABL1_.
Quantitative PCR assays were designed and used to re-measure MRD in remission bone marrow samples previously tested using Ig/TCR markers. All MRD testing complied with EuroMRD guidelines.
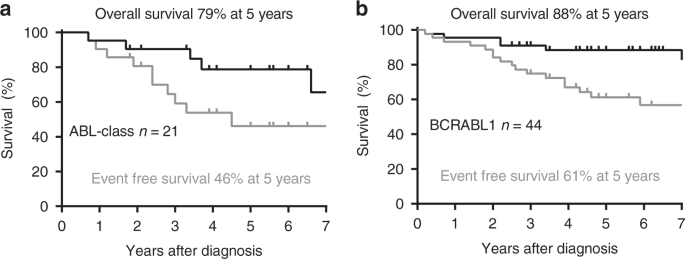
RESULTS ABL-class patients had 46% 5year event-free survival and 79% 5year overall survival. All had sensitive fusion tests giving high concordance between Ig/TCR and ABL-class fusion
results (21 patients, _n_ = 257 samples, r2 = 0.9786, _P_ < 0.0001) and Ig/TCR and _IKZF1_-deletion results (9 patients, _n_ = 143 samples, r2 = 0.9661, _P_ < 0.0001). In contrast, in
_BCR-ABL1_ patients, Ig/TCR and _BCR-ABL1_ tests were discordant in 32% (40 patients, _n_ = 346 samples, r2 = 0.4703, _P_ < 0.0001) and _IKZF1_-deletion results were closer to Ig/TCR (25
patients, _n_ = 176, r2 = 0.8631, _P_ < 0.0001). CONCLUSIONS MRD monitoring based on patient-specific assays detecting gene fusions or recurrent assays for _IKZF1_-deletions is feasible
and provides good alternatives to Ig/TCR tests to monitor MRD in ABL-class ALL. SIMILAR CONTENT BEING VIEWED BY OTHERS ‘EVALUATION OF ADVERSE PROGNOSTIC GENE ALTERATIONS & MRD POSITIVITY
IN _BCR::ABL1_-LIKE B-LINEAGE ACUTE LYMPHOBLASTIC LEUKAEMIA PATIENTS, IN A RESOURCE-CONSTRAINED SETTING Article 08 May 2023 ANALYSIS OF MEASURABLE RESIDUAL DISEASE BY IG/TR GENE
REARRANGEMENTS: QUALITY ASSURANCE AND UPDATED EUROMRD GUIDELINES Article Open access 14 May 2024 MINIMAL RESIDUAL DISEASE DETECTION BY NEXT-GENERATION SEQUENCING OF DIFFERENT IMMUNOGLOBULIN
GENE REARRANGEMENTS IN PEDIATRIC B-ALL Article Open access 17 November 2023 INTRODUCTION ABL-class fusions are a feature of approximately 3% of paediatric acute lymphoblastic leukaemia (ALL)
cases [1, 2] with similar gene expression patterns to Philadelphia chromosome positive (Ph-pos) ALL and with generally poor responses to standard induction chemotherapy. While Ph-pos ALL
results from a t(9;22) translocation creating a _BCR-ABL1_ fusion; this subset of Ph-like ALL cases involve the fusion of another gene expressed during lymphocyte differentiation such as
_EBF1_, _SSBP2_, _ETV6_, _NUP214_ with a gene encoding a tyrosine kinase or a receptor tyrosine kinase such as _PDGFRB_, _CSF1R_, _ABL1_ or _ABL2_. ALL patients with these ABL-class fusions
are generally sensitive to tyrosine kinase inhibitors (TKI) including dasatinib in vitro [3] and adjuvant TKIs in patients [4, 5]. Two recent studies from the AIEOP-BFM and Pont di Legno
groups showed the 5-year event-free survival (EFS) for patients with ABL-class fusions in the pre-TKI era was 49% (_n_ = 46) [1] and 59% (_n_ = 122) respectively [2]. The poor outcomes
associated with both _BCR-ABL1_ ALL and Ph-like ALL mean that many treating clinicians request close MRD monitoring for these patients, during initial therapy and particularly for
post-remission surveillance, including after HSCT. Moreover, in the TKI era, where HSCT is no longer indicated in many patients with _BCR-ABL1_ ALL [6], accurate determination of
post-treatment MRD is critical to identify patients with sub-optimal TKI response, where HSCT may still offer the best chance of cure. However, in a previous study of _BCR-ABL1_ ALL [7], we
demonstrated that MRD analysis using conventional MRD markers based on immunoglobulin and T-cell receptor (Ig/TCR) rearrangements fails to detect or underestimates MRD compared to qPCR
genomic tests detecting the _BCR-ABL1_ gene fusion itself in some CML-like patients. It is also accepted in _KMT2A_-rearranged infant ALL, that MRD testing based on detection of this disease
related fusion is not only feasible, but also preferable to Ig/TCR markers given the high incidence of oligoclonality and earlier stage of cell of origin that characterises ALL with
translocations of the KMT2A gene (previously known as MLL) [8, 9]. This collective knowledge raised the question of the reliability of immunoglobulin and T-cell receptor gene markers
(Ig/TCR) in patients with ABL-class fusions. Both _BCR-ABL1_ ALL and Ph-like ALL have a high incidence of _IKZF1_ deletions [10, 11]. Copy number analysis by microarray or by MLPA has
revealed a variety of _IKZF1_ deletions in ALL cases and their poor prognosis in newly diagnosed B-ALL was shown in German, Dutch, Italian and Australian cohorts [12,13,14,15]. International
collaborations have included _IKZF1_ deletions in multifactorial risk analysis [16, 17] and provided evidence that most if not all _IKZF1_ deletions are associated with high risk of relapse
[18] including recurrent internal deletions that are amenable to detection by generic qPCR MRD assays [19, 20]. These assays serve a dual purpose, with capacity to rapidly identify a subset
of _IKZF1_ high risk patients as well as to measure MRD without requiring prior sequencing. This study therefore set out to develop and evaluate patient-specific qPCR MRD assays for
paediatric ALL cases with _EBF1-PDGFRB, SSBP2-CSF1R_, _NUP214-ABL1_ and other _ABL1_ gene fusions and to compare the MRD results obtained with those based on _IKZF1_-deletion and
conventional Ig/TCR qPCR MRD measurements. We used two different next generation sequencing (NGS) strategies - targeted and whole genome sequencing - to determine the precise breakpoint
sequences needed to design patient-specific qPCR assays for ABL-class fusions. METHODS PATIENT SAMPLES This study was conducted on DNA samples from 65 paediatric ALL patients with parental
consent and human ethics approval. All bone marrow samples were originally tested for MRD in response to clinical requests and results reported in real-time. The same samples were stored for
research including retesting (with technical triplicates) to compare MRD levels obtained using alternate MRD assays. IDENTIFICATION OF PATIENTS WITH ABL-CLASS FUSIONS Patients with
ABL-class fusions were provisionally identified by several methods: (a) G-banded karyotyping and fluorescent in situ hybridization as performed as standard of care with _BCR, ABL1_ and
_PDGFRB_ at some centres; (b) MLPA analysis performed with SALSA P335 ALL-IKZF1 A4 or B1 kit (MRC-Holland, Amsterdam, the Netherlands) [15] since we discovered that patients with
_EBF1-PDFGRB_ have heterozygous loss of _EBF1_ exon 16; (c) Ph-like TLDA expression pattern in unselected cohort [21] or (d) patients referred on basis of high risk features (defined by
ANZCHOG 2014 guidelines). Fusion transcripts were analysed by RT-PCR and Sanger sequencing or RNA-Seq. Following provisional identification, precise genomic breakpoints were identified in
diagnostic DNA using multiplex long-distance PCR for _BCR-ABL1_ [7] or by analysis of targeted NGS for difficult _BCR-ABL1_ cases and 12 ABL-class cases. ANALYSIS OF BREAKPOINT SEQUENCE FOR
ABL-CLASS FUSIONS FROM WGS SEQUENCE Two of the ABL-class cases were enroled on the PRISM precision medicine trial (NCT03336931) which used WGS analysis to at least 90x-depth of leukaemia
cell DNA and 30x germline DNA [22]. The other seven ABL-class cases were sequenced using WGS to 30x coverage with no matched germline, reasoning that the somatic ABL-class fusions would be
readily identifiable. WGS was conducted at the Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research (Australia), using the Illumina HiSeq X Ten platform with a
paired-end read length of 150 bases. Sequencing libraries were prepared from more than 1 µg of DNA using KAPA PCR-Free v2.1 (Roche). Raw fastq files were aligned to the hs37d5 reference
genome using BWA-MEM (v0.17.10-r789) [23] with resulting BAM file reads marked using Novosort (v1.03.01; default settings). For cases with a matched germline, the WGS data were analysed as
previously described [22]. For cases without a matched germline, a tumour-only analysis pipeline was adopted, using the following steps: somatic SNVs and short indels (<50 bp) were
identified using Sage (v2.2) [24] and germline variants were filtered out using a panel of normals. The panel of normals contains variants identified from 1000 germline controls, sequenced
using WGS to 30–40x depth using HiSeq X10, where each variant was observed at least once, with at least three reads and a cumulative base quality of 30. Somatic variants were annotated using
SnpEff (v4_3t) [25] and imported into the in-house Glooee platform for filtration and prioritization. Tumour purity, ploidy and somatic copy number variants (CNVs) were identified using
PURPLE (v3.0) [24], and structural variants (SVs) were identified using GRIDSS (v2.9.4) [26] and then annotated using Ensembl genes. LINX (v1.16) was used to visualize SV clusters and
derivative chromosomes [27]. MRD Q-PCR ASSAYS TO DETECT ABL FUSIONS, IKZF1 DELETIONS AND IG/TCR REARRANGEMENTS MRD tests for ABL-class fusions involved patient specific primers and a Taqman
hydrolysis probe spanning the unique breakpoint sequence as reported previously for _BCR-ABL1_ [7] and testing bone marrow DNA samples usually retrospectively. The unique breakpoint
sequences for these patients and custom primers and probes are shown in Supplementary Tables 1 and 2. Routine PCR-MRD marker screening was performed by 24 single or multiplex PCR reactions
on leukaemic DNA to detect rearrangements in immunoglobulin heavy and kappa genes and T-cell receptor gamma, delta, beta and delta-alpha genes (Ig/TCR) followed by heteroduplex analysis and
direct Sanger sequencing. Unique breakpoint sequences were identified using the NCBI Nucleotide BLAST database, https://blast.ncbi.nlm.nih.gov/ or hardcopies circulated by the EuroMRD group.
The actual MRD testing involved q-PCR assays to detect these markers using one patient-specific primer and a gene segment specific primer and hydrolysis probe performed on an Biorad Icycler
or CFX platform in real time [28]. Diagnostic samples for all patients were screened for four _IKZF_1 MRD markers to detect internal gene deletions specifically _IKZF1_Δ2-7, _IKZF1_Δ4-7,
_IKZF1_Δ2-8, _IKZF1_Δ4-8, using generic qPCR tests with custom made primers and Locked Nucleic Acid (LNA) probes (Integrated DNA Technologies). For patients with a high level of marker
(>1 × 10−1 level present in positive controls), the same assay was then used with the patient’s own dilution curve to measure MRD in their remission samples. DATA ANALYSIS All MRD qPCR
tests, including the ABL-class fusion and _IKZF1_ deletion assays, were performed at the Children’s Cancer Institute and analysed according to the guidelines established by EuroMRD (van der
Velden et al, 2007). Standard curves met minimum standards of >0.98 correlation coefficient and slope between 3.1 and 3.9. MRD was scored positive or negative according to the definition
established for protocols in which therapy intensification is intended and with reference to normal peripheral blood mononuclear cell DNA samples. When comparing MRD levels determined by
different tests concordant results were defined as those with <1.0 log difference in quantitative results or either negative or non-quantifiable results with both marker tests. In
contrast discordant results had >1.0 log difference in quantifiable results. When one marker gave a quantifiable result and the other gave non-quantifiable or negative result, the
quantitative range was considered. Results were defined as discordant if there was <1.0 log difference between the quantitative result and the quantitative range for the assay used for
non-quantifiable positive or negative result. Survival times were measured from date of diagnosis to date of relapse or death, or to last clinic visit for patients without these events.
Kaplan-Meier survival curves were generated and log-rank tests applied using GraphPad Prism version 7.04. RESULTS DESCRIPTION OF PATIENTS WITH ABL-CLASS FUSIONS AND _BCR-ABL1_ A database
search of ALL patients with both qPCR MRD testing and research consent identified 21 paediatric patients with ABL-class fusions including 19 B-ALL and two T-ALL (Table 1). These patients
were diagnosed between 2004 and 2018 with a median follow up of 5.5 years for survivors. We also identified an additional 44 children with _BCR-ABL1_ ALL diagnosed in that timeframe (median
follow up of 5.7 years) for comparison. The most frequently identified ABL-class fusions were _EBF1-PDGFRB_ (9 cases, 43%) and _NUP214-ABL1_ (4 cases, 19%). Two patients had _SSBP2-CSF1R_,
two had other _PDGFRB_ fusions and the remaining four patients had other _ABL1_ fusions. The patients were predominantly male (71%) with median age of 10 years and 82% had high MRD at end of
induction. In contrast to the others, the two patients with _ETV6-ABL1_ fusion were both one year old females with complete MRD response (MRD negative) (Table 1). As a group, the ABL-class
fusions had 5-year EFS of 46% and 5-year OS of 79%. While it appears from the Fig. 1a and b data, that outcomes may be poorer for patients with ABL-class compared to _BCR-ABL1_, these
differences were not statistically significant. It is also worth noting that seven (33%) of the ABL-class patients did not receive a TKI (imatinib or dasatinib) in first remission, compared
to one of 44 _BCR-ABL1_ patients (Table 1). COMPARISON OF ABL-CLASS FUSION MRD TESTS WITH IG/TCR TESTS In order to determine precise genomic breakpoints and to design fusion-detecting qPCR
MRD assays, targeted next generation sequencing (NGS) was performed on diagnostic bone marrow DNA from the 44 _BCR-ABL1_ cases and 12 of the ABL-class patients. These included all cases with
fusions involving _ABL1_, except one _NUP214-ABL1_, and five cases with _PDGFRB_ fusions. For the remaining nine patients, whole genome sequencing (WGS) was performed and analysed for
breakpoints including five with _EBF1-PDGFRB_, 1 _ATP7IP-PDGFRB_, 1 _NUP214-ABL1_ and 2 _SSBP2-CSF1R_. These breakpoints were used to design patient-specific qPCR assays composed of two
primers specific for each of the genes involved and a hydrolysis probe spanning the precise breakpoint sequence. Each assay was then evaluated for quantitative range and sensitivity
according to the widely used EuroMRD guidelines that evaluate amplification of duplicates in a standard curve created from a dilution series of the diagnostic bone marrow DNA sample and 6
normal DNA samples from mononuclear cells. The analysis in Fig. 2a showed that the assays measuring the ABL-class fusion breakpoints had adequate sensitivity (1 × 10-4) in all and were
highly sensitive in most patients (1 × 10-5 in 81%), with an acceptable quantitative range (QR at least 1 × 10-4) for all patients except one with QR of 5 × 10-4 and superior quantitation in
71% (QR of 5 × 10-5). These standardised MRD assay metrics compared favourably with conventional Ig/TCR based MRD tests in the same patients and same samples tested earlier and reported in
diagnostic MRD reports (Fig. 2a). These new MRD assays were then utilized to re-measure MRD levels in 257 bone marrow samples from the 21 ABL-class patients previously tested using Ig/TCR
marker tests (Fig. 3a). The MRD data obtained were highly correlated with Ig/TCR results (Pearson correlation coefficient r2 of 0.9123, _P_ < 0.0001). All of the patients had concordant
results, defined by <1.0 log difference in quantifiable results or non-quantifiable or negative results for both samples. A similar comparison was performed for 40 _BCR-ABL1_ patients
whose MRD had been evaluated with MRD assays designed to detect the genomic breakpoint as well as an Ig/TCR clonal marker (Fig. 3b). In contrast to ABL-class patients, 13 _BCR-ABL1_ patients
(33%) had discordant MRD results with higher MRD results obtained with the _BCR-ABL1_ marker in 12 cases and higher Ig/TCR MRD levels in the remaining patient. These marked differences in
some patients contributed to a much lower correlation coefficient for the results of the parallel MRD testing of the same samples using 2 different marker types in _BCR-ABL1_ patients (40
cases, r2 = 0.4703, _P_ < 0.0001, Fig. 3b). COMPARISON OF _IKZF_1 MRD TESTS WITH IG/TCR TESTS All ABL-class and _BCR-ABL1_ patients were also screened with four qPCR assays designed to
detect _IKZF_1Δ4-7, _IKZF_1Δ2-7, _IKZF_1Δ2-8 and _IKZF_1Δ4-8 respectively (Fig. 4). Of the 21 ABL-class fusion cases, nine (43%) showed a high level of at least one of these deletions gauged
to be suitable for an effective MRD test because the level of deletion was greater than 1 × 10-1 compared to our positive control DNA for the deletion derived from primary patient cells or
patient-derived xenograft. An additional two cases had a deletion at a sub-clonal level (Table 1). In comparison, screening for the four _IKZF_1 deletion assays identified 26 (59%) of the
_BCR-ABL1_ patients with a high level for one of these dual-purpose markers. The sensitivity and QR levels for these generic _IKZF_1 qPCR tests were assessed using a dilution series of each
patient’s diagnostic DNA to create standard curves that were also used to measure the remission samples in the same assay. The sensitivity and QR levels for generic _IKZF_1 assays were
similar to the Ig/TCR assays for both ALL subtypes (Fig. 2a, b). When the MRD levels measured by _IKZF_1 markers were compared with the results of Ig/TCR assays for the same 143 samples from
ABL-class patients there was a > 1 log difference for only a single sample (from Patient 14) Fig. 3c. For _BCR-ABL1_ patients, three patients had discordant results with samples giving
different MRD levels (>1 log difference). The overall correlation for MRD levels measured by the _IKZF_1 compared with Ig/TCR markers in ABL-class patients were better for patients with
ABL-class fusions (r2 = 0.9661, Fig. 3c) than _BCR-ABL1_ (r2 = 0.8631, Fig. 3d). COMPARISON OF _IKZF1_ MRD TESTS WITH FUSION MRD TESTS To improve our understanding of the variations in MRD
level that are sometimes observed with different markers in particular patients, further comparisons were made between the MRD results obtained with the fusion-based versus _IKZF_1
deletion-based markers (Fig. 3e, f). The results of these two types of MRD tests on the same samples from patients with _BCR-ABL1_ fusions showed relatively low Pearson’s correlation
coefficient (0.5386, Fig. 3f). This reflects the observation that at the patient level, 6/26 (23%) patients had discordant MRD results. We were able to compare MRD determined by all three
marker types for 23 _BCR-ABL1_ patients –16 patients (70%) were concordant in all samples. Again, higher concordance was observed for cases with ABL-class fusions compared to _BCR-ABL1_
patients with none of the nine ABL-class patients showing discordant results with _IKZF1_ markers (>1 log difference). In fact only 3 of 143 comparisons showed a difference >0.5 log
(the 3 all from Patient 14) leading to a high Pearson’s coefficient (Fig. 3e, r2 = 0.9751, _P_ < 0.0001). DISCUSSION This study demonstrated the feasibility of using targeted NGS or WGS
to define genomic breakpoints and to design satisfactory qPCR MRD assays to monitor disease in ALL patients with ABL-class fusions. These 21 patients had poor outcomes, comparable to larger
studies [1, 2], although we would expect this to improve with earlier diagnosis of the fusions and consistent intervention with TKIs [5]. The study also found a high incidence of recurrent
_IKZF1_ deletions in ABL-class patients consistent with a previous report of 40% _IKZF1_ deletions in Ph-like ALL [11] and demonstrated effective use of these markers to monitor disease.
Both the fusion and deletion MRD tests showed generally highly comparable results versus conventional Ig/TCR markers, thus providing additional scope to monitor MRD in these patients. MRD
tests based on _IKZF1_-deletions were of particular interest since these deletions occurred in 23% of relapsed B-ALL [29] as well as Ph-like ALL and approximately half are recurrent and can
be detected in multiple patients by generic assays [19, 20]. In this study, it was feasible to use these dual-purpose markers which do not require any prior sequencing to perform MRD testing
in 43% of ABL-class patients and 59% of _BCR-ABL1_ patients. The analyses in this paper complement previous studies showing the potential to use genomic breakpoints for _BCR-ABL1_ [7];
_KMT2A_-rearrangements [8, 9]; and _CDKN2A/B_ deletions [30] as MRD markers in ALL. This is in line with our hypothesis that disease-drivers will make more reliable MRD markers than
disease-passengers such as clonal Ig/TCR markers. While Ig/TCR markers have served as effective MRD markers for ALL, particularly when two markers are used as recommended by most trials
[31], their use is restricted to lymphoid disease, they can underestimate MRD in _KMT2A_-rearranged infant ALL [9] and they can be subject to clonal evolution or selection at relapse [32].
However, each new genomic fusion or deletion marker should be considered on its own merits by careful evaluation and comparison with other methodologies before general diagnostic use for ALL
or other diseases. In our laboratory, recurrent ALL genomic deletions in _BTLA_ and _SLX4IP_ genes did not appear to provide stable MRD markers (unpublished data). The approach used herein
has the advantage that the well-established EuroMRD guidelines for evaluating the quantitative range and sensitivity of MRD tests in ALL can be readily applied to new genomic breakpoint
assays and their MRD results, allowing fair comparisons to be made with measurement of Ig/TCR markers by either q-PCR [33] or by ddPCR [30]. Monitoring residual disease in patients with
_BCR-ABL1_ is challenging. Our previous study identified that a subset of ALL patients with _BCR-ABL1_ and CML-like features had discordant MRD results measured by qRT-PCR or qPCR for
_BCR-ABL1_ and Ig/TCR [7]. This study used an extended series of _BCR-ABL1_ patients and found discordancy in qPCR results of >1 log difference in 33% of patients between BCR-ABL1 and
Ig/TCR compared to none of the 21 ABL-class ALL cases. The additional use of qPCR _IKZF1_ deletion markers available in 59% of the _BCR-ABL1_ patients did not progress our understanding
although it served to illustrate further the inherent clonal instability in some of these patients. We have no definitive answer on the best way to monitor residual disease in _BCR-ABL1_ ALL
patients although the results of on-going trials such as EsPhALL trial will be informative. Finally, as whole genome sequencing becomes more commonly used for ALL patients, this study shows
it provides a viable alternative to multiple PCRs followed by heteroduplex analysis and Sanger sequencing for the detection of MRD markers in ALL. The ability to define the precise
patient-specific genomic breakpoint sequences for key disease-related fusions and deletions that drive disease also suggests significant opportunity to develop in liquid biopsy assays for
MRD in other cancers [30]. DATA AVAILABILITY Data are securely stored at Children’s Cancer Institute with access limited by ethical considerations. REFERENCES * Cario G, Leoni V, Conter V,
Attarbaschi A, Zaliova M, Sramkova L, et al. Relapses and treatment-related events contributed equally to poor prognosis in children with ABL-class fusion positive B-cell acute lymphoblastic
leukemia treated according to AIEOP-BFM protocols. Haematologica. 2020;105:1887–94. Article CAS Google Scholar * den Boer ML, Cario G, Moorman AV, Boer JM, de Groot-Kruseman HA, Fiocco
M, et al. Outcomes of paediatric patients with B-cell acute lymphocytic leukaemia with ABL-class fusion in the pre-tyrosine-kinase inhibitor era: a multicentre, retrospective, cohort study.
Lancet Haematol. 2021;8:e55–e66. Article Google Scholar * Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute
lymphoblastic leukemia. N Engl J Med. 2014;371:1005–15. Article Google Scholar * Tanasi I, Ba I, Sirvent N, Braun T, Cuccuini W, Ballerini P, et al. Efficacy of tyrosine kinase inhibitors
in Ph-like acute lymphoblastic leukemia harboring ABL-class rearrangements. Blood. 2019;134:1351–5. Article Google Scholar * Moorman AV, Schwab C, Winterman E, Hancock J, Castleton A,
Cummins M, et al. Adjuvant tyrosine kinase inhibitor therapy improves outcome for children and adolescents with acute lymphoblastic leukaemia who have an ABL-class fusion. Br J Haematol.
2020;191:844–51. Article CAS Google Scholar * Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB, et al. Long-term follow-up of imatinib in pediatric Philadelphia
chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group Study AALL0031. Leukemia. 2014;28:1467–71. Article CAS Google Scholar * Hovorkova L, Zaliova M, Venn NC,
Bleckmann K, Trkova M, Potuckova E, et al. Monitoring of childhood ALL using BCR-ABL1 genomic breakpoints identifies a subgroup with CML-like biology. Blood. 2017;129:2771–81. Article CAS
Google Scholar * Burmeister T, Marschalek R, Schneider B, Meyer C, Gokbuget N, Schwartz S, et al. Monitoring minimal residual disease by quantification of genomic chromosomal breakpoint
sequences in acute leukemias with MLL aberrations. Leukemia. 2006;20:451–7. Article CAS Google Scholar * Van der Velden VH, Corral L, Valsecchi MG, Jansen MW, De Lorenzo P, Cazzaniga G,
et al. Prognostic significance of minimal residual disease in infants with acute lymphoblastic leukemia treated within the Interfant-99 protocol. Leukemia. 2009;23:1073–9. Article Google
Scholar * Mullighan CG, Su X, Zhang J, Radtke I, Phillips LAA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80. Article CAS
Google Scholar * van der Veer A, Waanders E, Pieters R, Willemse ME, Van Reijmersdal SV, Russell LJ, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but
not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122:2622–9. Article Google Scholar * Kuiper RP, Waanders E, van der Velden VHJ, van Reijmersdal SV,
Venkatachalam R, Scheijen B, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–64. Article CAS Google Scholar * Dorge P,
Meissner B, Zimmermann M, Moricke A, Schrauder A, Bouquin JP, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the
ALL-BFM 2000 protocol. Haematologica. 2013;98:428–32. Article Google Scholar * Palmi C, Valsecchi MG, Longinotti G, Silvestri D, Carrino V, Conter V, et al. What is the relevance of Ikaros
gene deletions as a prognostic marker in pediatric Philadelphia-negative B-cell precursor acute lymphoblastic leukemia? Haematologica. 2013;98:1226–31. Article CAS Google Scholar *
Sutton R, Venn NC, Law T, Boer JM, Trahair TN, Ng A, et al. A risk score including microdeletions improves relapse prediction for standard and medium risk precursor B-cell acute
lymphoblastic leukaemia in children. Br J Haematol. 2018;180:550–62. Article CAS Google Scholar * Hamadeh L, Enshaei A, Schwab C, Alonso CN, Attarbaschi A, Barbany G, et al. Validation of
the United Kingdom copy-number alteration classifier in 3239 children with B-cell precursor ALL. Blood Adv. 2019;3:148–57. Article CAS Google Scholar * Stanulla M, Cave H, Moorman AV.
IKZF1 deletions in pediatric acute lymphoblastic leukemia: still a poor prognostic marker? Blood. 2020;135:252–60. Article CAS Google Scholar * Boer JM, van der Veer A, Rizopoulos D,
Fiocco M, Sonneveld E, de Groot-Kruseman HA, et al. Prognostic value of rare IKZF1 deletion in childhood B-cell precursor acute lymphoblastic leukemia: an international collaborative study.
Leukemia. 2016;30:32–8. Article CAS Google Scholar * Venn NC, van der Velden VH, de Bie M, Waanders E, Giles JE, Law T, et al. Highly sensitive MRD tests for ALL based on the IKZF1
Delta3-6 microdeletion. Leukemia. 2012;26:1414–6. Article CAS Google Scholar * Caye A, Beldjord K, Mass-Malo K, Drunat S, Soulier J, Gandemer V, et al. Breakpoint-specific multiplex
polymerase chain reaction allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia. Haematologica.
2013;98:597–601. Article CAS Google Scholar * Heatley SL, Sadras T, Kok CH, Nievergall E, Quek K, Dang P, et al. High prevalence of relapse in children with Philadelphia-like acute
lymphoblastic leukemia despite risk-adapted treatment. Haematologica. 2017;102:e490–e3. Article CAS Google Scholar * Wong M, Mayoh C, Lau LMS, Khuong-Quang DA, Pinese M, Kumar A, et al.
Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat Med. 2020;26:1742–53. Article CAS Google Scholar * Li H.
Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv. 2013; https://arxiv.org/abs/1303.3997. * Cameron DL, Baber J, Shale C, Papenfuss AT,
Valle-Inclan JE, Besselink N, et al. GRIDSS, PURPLE, LINX: unscrambling the tumor genome via integrated analysis of structural variation and copy number. Preprint at bioRxiv. 2019;
https://www.biorxiv.org/content/10.1101/781013v1. * Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single
nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. Article CAS Google Scholar * Cameron DL, Schroder J,
Penington JS, Do H, Molania R, Dobrovic A, et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017;27:2050–60.
Article CAS Google Scholar * Shale C, Baber C, Cameron DL, Wong M, Cowley MJ, Papenfuss AT, et al. Unscrambling cancer genomes via integrated analysis of structural variation and copy
number. Preprint for BioRxiv. 2020; https://www.biorxiv.org/content/10.1101/2020.12.03.410860v1. * Sutton R, Shaw PJ, Venn NC, Law T, Dissanayake A, Kilo T, et al. Persistent MRD before and
after allogeneic BMT predicts relapse in children with acute lymphoblastic leukaemia. Br J Haematol. 2015;168:395–404. Article Google Scholar * Irving JA, Enshaei A, Parker CA, Sutton R,
Kuiper RP, Erhorn A, et al. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood.
2016;128:911–22. Article CAS Google Scholar * Subhash VV, Huang L, Kamili A, Wong M, Chen D, Venn NC, et al. Whole genome sequencing facilitates patient-specific quantitative PCR-based
minimal residual disease monitoring in acute lymphoblastic leukaemia, neuroblastoma and Ewing sarcoma. British J Cancer. 2022;126;482–91. * Flohr T, Schrauder A, Cazzaniga G, Panzer-Grümayer
R, van der Velden V, Fischer S, et al. Minimal residual disease-directed risk stratification using real-time quantitative PCR analysis of immunoglobulin and T-cell receptor gene
rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia. Leukemia. 2008;22:771–82. Article CAS Google Scholar * Choi S,
Henderson M, Kwan EN, Sutton R, Giles J, Venn NC, et al. Relapse in children with acute lymphoblastic leukaemia involving selection of a pre-existing drug resistant subclone. Blood.
2007;110:632–9. Article CAS Google Scholar * van der Velden V, Cazzaniga G, Schrauder A, Hancock JF, Bader P, Panzer E, et al. Analysis of minimal residual disease by Ig/TCR gene
rearrangements: Guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21:604–11. Article Google Scholar Download references ACKNOWLEDGEMENTS We thank Emma
McCormack for assisting with ethics applications; staff of the Children’s Cancer Institute Tumour Bank and Queensland Cell and Tissue bank for sample processing and banking and Erika Ong,
Amy Smalley and Lynda Saunders who assisted in data collection. Children’s Cancer Institute is affiliated with Sydney Children’s Hospital and UNSW. FUNDING This study was supported by Cancer
Australia PdCCRS1128727 funding awarded to RS, TNT, PS and DLW which provided salary for TL and supported MRD testing, RNA and WGS sequencing. The Czech Health Research Council
NU21-03-00128 funding (CI Jan Zuna) supported targeted breakpoint analysis. Salary for LHu was provided by The Cancer Council NSW Program Grant (CI Murray Norris) and Kids Cancer Alliance
grant (CI Mark Cowley). Open Access funding enabled and organized by CAUL and its Member Institutions. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Molecular Diagnostics, Children’s Cancer
Institute, Lowy Cancer Research Centre, UNSW, Sydney, NSW, Australia Nicola C. Venn, Libby Huang, Walter Muskovic, Tamara Law, Katerina Bendak, Murray D. Norris, Michelle J. Henderson, Toby
N. Trahair & Rosemary Sutton * Department of Paediatric Haematology and Oncology, Second Faculty of Medicine, Charles University, Prague, Czech Republic Lenka Hovorková & Jan Zuna *
CLIP—Childhood Leukaemia Investigation Prague, Prague, Czech Republic Lenka Hovorková & Jan Zuna * Computational Biology, Children’s Cancer Institute, Lowy Cancer Research Centre, UNSW,
Sydney, NSW, Australia Marie Wong & Mark J. Cowley * Precision Medicine Theme, South Australian Health and Medical Research Institute, Adelaide, SA, Australia Susan L. Heatley &
Deborah L. White * School of Medicine, University of Adelaide, Adelaide, SA, Australia Susan L. Heatley, Tom Revesz & Deborah L. White * Children’s Cancer Centre, The Royal Children’s
Hospital, Melbourne, VIC, Australia Seong Lin Khaw * Department of Clinical Haematology and Oncology, Women’s and Children’s Hospital, Adelaide, SA, Australia Tom Revesz * Cancer Centre for
Children, The Children’s Hospital at Westmead, Sydney, NSW, Australia Luciano Dalla Pozza & Peter J. Shaw * Blood and Bone Marrow Transplant Program, Queensland Children’s Hospital,
Brisbane, QLD, Australia Chris Fraser * Paediatric Oncology, Queensland Children’s Hospital, Brisbane, QLD, Australia Andrew S. Moore * Children’s Haematology/Oncology Centre Christchurch
Hospital, Christchurch, New Zealand Siobhan Cross * School of Women’s and Children’s Health, University of New South Wales, Sydney, NSW, Australia Murray D. Norris, Michelle J. Henderson,
Mark J. Cowley, Toby N. Trahair & Rosemary Sutton * Kids Cancer Centre, Sydney Children’s Hospital, Randwick, NSW, Australia Toby N. Trahair * University Hospital Motol, Prague, Czech
Republic Jan Zuna Authors * Nicola C. Venn View author publications You can also search for this author inPubMed Google Scholar * Libby Huang View author publications You can also search for
this author inPubMed Google Scholar * Lenka Hovorková View author publications You can also search for this author inPubMed Google Scholar * Walter Muskovic View author publications You can
also search for this author inPubMed Google Scholar * Marie Wong View author publications You can also search for this author inPubMed Google Scholar * Tamara Law View author publications
You can also search for this author inPubMed Google Scholar * Susan L. Heatley View author publications You can also search for this author inPubMed Google Scholar * Seong Lin Khaw View
author publications You can also search for this author inPubMed Google Scholar * Tom Revesz View author publications You can also search for this author inPubMed Google Scholar * Luciano
Dalla Pozza View author publications You can also search for this author inPubMed Google Scholar * Peter J. Shaw View author publications You can also search for this author inPubMed Google
Scholar * Chris Fraser View author publications You can also search for this author inPubMed Google Scholar * Andrew S. Moore View author publications You can also search for this author
inPubMed Google Scholar * Siobhan Cross View author publications You can also search for this author inPubMed Google Scholar * Katerina Bendak View author publications You can also search
for this author inPubMed Google Scholar * Murray D. Norris View author publications You can also search for this author inPubMed Google Scholar * Michelle J. Henderson View author
publications You can also search for this author inPubMed Google Scholar * Deborah L. White View author publications You can also search for this author inPubMed Google Scholar * Mark J.
Cowley View author publications You can also search for this author inPubMed Google Scholar * Toby N. Trahair View author publications You can also search for this author inPubMed Google
Scholar * Jan Zuna View author publications You can also search for this author inPubMed Google Scholar * Rosemary Sutton View author publications You can also search for this author
inPubMed Google Scholar CONTRIBUTIONS RS, TNT, and JZ designed the study, obtained ethics and wrote the manuscript. All authors reviewed and approved the manuscript. RS, MDN, DLW, PJS, TNT,
MJC, JZ obtained essential research grants. NCV, LHu, WM, TL designed qPCR assays and performed MRD tests. RS, MH, TL assessed and reported MRD tests. LH and JZ performed targeted NGS and
breakpoint analyses. ME and MJC designed and performed WGS analyses. NCV, SH and DLW performed MLPA and RNA seq and KB analysed sequences. SLK, TR, LDP, PJS, CF, ASM, SC and TNT all provided
clinical data. CORRESPONDING AUTHOR Correspondence to Rosemary Sutton. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ETHICS APPROVAL AND CONSENT TO
PARTICIPATE This study was conducted in accordance with the Declaration of Helsinki and the NHMRC Australia National Statement on Ethical Conduct in Human Research (2007). It used leftover
samples from paediatric ALL patients, all with signed parental consent for MRD testing and future research. approved Both the research on new MRD markers to improve stratification
(2019/ETH06161) and the parental information and consent forms (LNR/13/SCHN/392) were approved by the Sydney Children’s Hospital Network Human Research Ethics Committee. CONSENT FOR
PUBLICATION Parents of all patients had signed consent for publication of future research in a format that does not identify the patients. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer
Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLE 1 SUPPLEMENTARY TABLE 2 CHECKLIST
RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and
reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes
were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If
material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain
permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Venn, N.C., Huang, L., Hovorková, L. _et al._ Measurable residual disease analysis in paediatric acute lymphoblastic leukaemia patients with ABL-class fusions. _Br J Cancer_ 127,
908–915 (2022). https://doi.org/10.1038/s41416-022-01806-6 Download citation * Received: 17 September 2021 * Revised: 12 March 2022 * Accepted: 25 March 2022 * Published: 01 June 2022 *
Issue Date: 01 September 2022 * DOI: https://doi.org/10.1038/s41416-022-01806-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable
link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
