Consensus molecular subtyping of metastatic colorectal cancer expands biomarker-directed therapeutic benefit for patients with cms1 and cms2 tumors
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT BACKGROUND We developed a whole transcriptome sequencing (WTS)-based Consensus Molecular Subtypes (CMS) classifier using FFPE tissue and investigated its prognostic and predictive
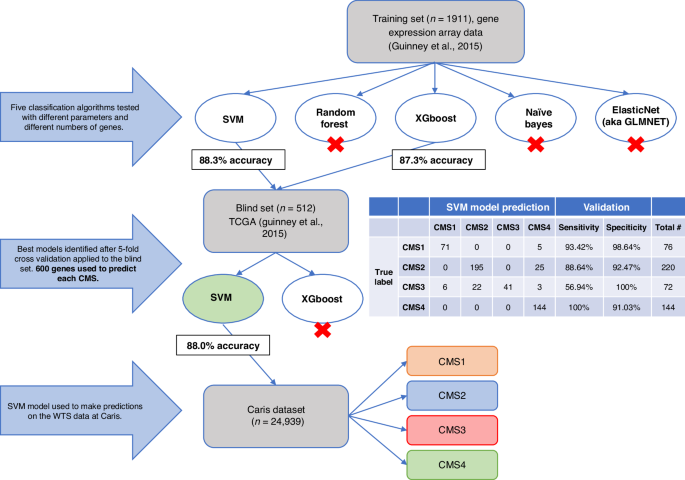
utility in a large clinico-genomic database of CRC patients (_n_ = 24,939). METHODS The classifier was trained against the original CMS datasets using an SVM model and validated in an
independent blinded TCGA dataset (88.0% accuracy). Kaplan–Meier estimates of overall survival (OS) and time-on-treatment (TOT) were calculated for each CMS (_p_ < 0.05 considered
significant). RESULTS CMS2 tumors were enriched on left-side of colon and conferred the longest median OS. In _RAS_-wildtype mCRC, left-sided tumors and CMS2 classification were associated
with longer TOT with anti-EGFR antibodies (cetuximab and panitumumab). When restricting to only CMS2, there was no significant difference in TOT between right- versus left-sided tumors. CMS1
tumors were associated with a longer median TOT with pembrolizumab relative to other CMS groups, even when analyzing only microsatellite stable (MSS) tumors. DISCUSSION A WTS-based CMS
classifier allowed investigation of a large multi-institutional clinico-genomic mCRC cohort, suggesting anti-EGFR therapy benefit for right-sided _RAS_-WT CMS2 tumors and immune checkpoint
inhibitor benefit for MSS CMS1. Routine CMS classification of CRC provides important treatment associations that should be further investigated. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 24 print issues and online
access $259.00 per year only $10.79 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which
are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
TARGETED THERAPY GUIDED BY CIRCULATING TUMOR DNA ANALYSIS IN ADVANCED GASTROINTESTINAL TUMORS Article Open access 16 September 2024 COMPREHENSIVE ASSESSMENT OF ACTIONABLE GENOMIC
ALTERATIONS IN PRIMARY COLORECTAL CARCINOMA USING TARGETED NEXT-GENERATION SEQUENCING Article Open access 16 July 2022 WHOLE-EXOME TUMOR-AGNOSTIC CTDNA ANALYSIS ENHANCES MINIMAL RESIDUAL
DISEASE DETECTION AND REVEALS RELAPSE MECHANISMS IN LOCALIZED COLON CANCER Article Open access 29 April 2025 DATA AVAILABILITY The data generated in this study are not publicly available due
to data size and patient privacy but summarized data are available upon reasonable request from the corresponding author. REFERENCES * Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A.
Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73:233–54. Article PubMed Google Scholar * Atreya CE, Yaeger R, Chu E. Systemic Therapy for Metastatic Colorectal Cancer: From
Current Standards to Future Molecular Targeted Approaches. Am Soc Clin Oncol Educ Book. 2017;37:246–56. Article PubMed Google Scholar * Weng J, Li S, Zhu Z, Liu Q, Zhang R, Yang Y, et al.
Exploring immunotherapy in colorectal cancer. J Hematol Oncol. 2022;15:95. Article CAS PubMed PubMed Central Google Scholar * Zeineddine FA, Zeineddine MA, Yousef A, Gu Y, Chowdhury S,
Dasari A, et al. Survival improvement for patients with metastatic colorectal cancer over twenty years. npj Precis Oncol. 2023;7:16. Article CAS PubMed PubMed Central Google Scholar *
Roepman P, Schlicker A, Tabernero J, Majewski I, Tian S, Moreno V, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and
epithelial-to-mesenchymal transition. Int J Cancer. 2014;134:552–62. Article CAS PubMed Google Scholar * Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, et al. Gene
expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231:63–76. Article CAS PubMed PubMed Central Google Scholar * Schlicker A, Beran
G, Chresta CM, McWalter G, Pritchard A, Weston S, et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics.
2012;5:66. Article CAS PubMed PubMed Central Google Scholar * Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification
system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–25. Article CAS PubMed PubMed Central Google Scholar * De Sousa EMF, Wang X, Jansen M, Fessler
E, Trinh A, de Rooij LP, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614–8. Article Google
Scholar * Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and
prognostic value. PLoS Med. 2013;10:e1001453. Article CAS PubMed PubMed Central Google Scholar * Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The
consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. Article CAS PubMed PubMed Central Google Scholar * Lenz HJ, Ou FS, Venook AP, Hochster HS, Niedzwiecki D,
Goldberg RM, et al. Impact of Consensus Molecular Subtype on Survival in Patients With Metastatic Colorectal Cancer: Results From CALGB/SWOG 80405 (Alliance). J Clin Oncol. 2019;37:1876–85.
Article PubMed PubMed Central Google Scholar * Stintzing S, Wirapati P, Lenz HJ, Neureiter D, Fischer von Weikersthal L, Decker T, et al. Consensus molecular subgroups (CMS) of
colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann Oncol. 2019;30:1796–803. Article CAS PubMed PubMed Central
Google Scholar * Mooi JK, Wirapati P, Asher R, Lee CK, Savas P, Price TJ, et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab
benefit in metastatic colorectal cancer: molecular analysis of the AGITG MAX clinical trial. Ann Oncol. 2018;29:2240–6. Article CAS PubMed Google Scholar * Borelli B, Fontana E, Giordano
M, Antoniotti C, Lonardi S, Bergamo F, et al. Prognostic and predictive impact of consensus molecular subtypes and CRCAssigner classifications in metastatic colorectal cancer: a
translational analysis of the TRIBE2 study. ESMO Open. 2021;6:100073. Article CAS PubMed PubMed Central Google Scholar * Stahler A, Hoppe B, Na IK, Keilholz L, Müller L, Karthaus M, et
al. Consensus Molecular Subtypes as Biomarkers of Fluorouracil and Folinic Acid Maintenance Therapy With or Without Panitumumab in RAS Wild-Type Metastatic Colorectal Cancer (PanaMa, AIO KRK
0212). J Clin Oncol. 2023;41:2975–87. Article CAS PubMed Google Scholar * Okita A, Takahashi S, Ouchi K, Inoue M, Watanabe M, Endo M, et al. Consensus molecular subtypes classification
of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9:18698–711. Article PubMed PubMed Central Google Scholar
* Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite
Instability-High Metastatic Colorectal Cancer. J Clin Oncol. 2018;36:773–9. Article CAS PubMed Google Scholar * Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1
blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. Article CAS PubMed Google Scholar * Dienstmann R, Salazar R, Tabernero J. Overcoming Resistance to
Anti-EGFR Therapy in Colorectal Cancer. Am Soc Clin Oncol Educ Book. 2015;35:e149–e56. * Benson AB 3rd, Venook AP, Cederquist L, Chan E, Chen YJ, et al. Colon Cancer, Version 1.2017, NCCN
Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2017;15:370–98. Article CAS PubMed Google Scholar * Bahl A, Talwar V, Sirohi B, Mehta P, Arya D, Shrivastava G, et al.
Primary Tumor Location as a Prognostic and Predictive Marker in Metastatic Colorectal Cancer (mCRC). Front Oncol. 2020;10:964. Article PubMed PubMed Central Google Scholar * Chowdhury S,
Gupta R, Millstein J, Lin K, Haridas V, Zeineddine MA, et al. Transcriptional Profiling and Consensus Molecular Subtype Assignment to Understand Response and Resistance to Anti-Epidermal
Growth Factor Receptor Therapy in Colorectal Cancer. JCO Precis Oncol. 2023;7:e2200422. Article PubMed PubMed Central Google Scholar * Shaikh H, McGrath JE, Hughes B, Xiu J, Brodskiy P,
Sukari A, et al. Genomic and Molecular Profiling of Human Papillomavirus Associated Head and Neck Squamous Cell Carcinoma Treated with Immune Checkpoint Blockade Compared to Survival
Outcomes. Cancers. 2021;13:6309. Article CAS PubMed PubMed Central Google Scholar * Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour
mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study.
Lancet Oncol. 2020;21:1353–65. Article CAS PubMed Google Scholar * Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by
next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7:746–56. Article CAS PubMed PubMed Central Google Scholar * Dobin A,
Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. Article CAS PubMed Google Scholar * Patro R,
Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9. Article CAS PubMed PubMed Central
Google Scholar * Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science.
2018;362:eaar3593. Article PubMed PubMed Central Google Scholar * Gomez OH, Soto VH, Machado I, Mendez MC, Cuatrecasas M, Horndler C, et al. 474P Prognostic and predictive role of
Consensus Molecular Subtypes (CMS) determined by immunohistochemistry in metastatic colorectal cancer (mCRC). Ann Oncol. 2020;31:S442–S3. Article Google Scholar * Purcell RV, Schmeier S,
Lau YC, Pearson JF, Frizelle FA. Molecular subtyping improves prognostication of Stage 2 colorectal cancer. BMC Cancer. 2019;19:1155. Article CAS PubMed PubMed Central Google Scholar *
Venook AP, Niedzwiecki D, Innocenti F, Fruth B, Greene C, O’Neil BH, et al. Impact of primary (1°) tumor location on overall survival (OS) and progression-free survival (PFS) in patients
(pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405 (Alliance). J Clin Oncol. 2016;34:3504. Article Google Scholar * Cremolini C, Antoniotti C, Moretto R, Masi G,
Falcone A. First-line therapy for mCRC — the influence of primary tumour location on the therapeutic algorithm. Nat Rev Clin Oncol. 2017;14:113. Article PubMed Google Scholar * Holch JW,
Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. Eur J
Cancer. 2017;70:87–98. Article PubMed Google Scholar * Fiala O, Ostasov P, Hosek P, Sorejs O, Liska V, Buchler T, et al. The Predictive Role of Primary Tumour Sidedness in Metastatic
Colorectal Cancer Treated With Targeted Agents. Anticancer Res. 2019;39:5645–52. Article CAS PubMed Google Scholar * Arnold D, Lueza B, Douillard JY, Peeters M, Lenz HJ, Venook A, et al.
Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized
trials. Ann Oncol. 2017;28:1713–29. Article CAS PubMed PubMed Central Google Scholar * Brulé SY, Jonker DJ, Karapetis CS, O’Callaghan CJ, Moore MJ, Wong R, et al. Location of colon
cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur J Cancer. 2015;51:1405–14. Article PubMed Google Scholar *
Ciardiello D, Vitiello PP, Cardone C, Martini G, Troiani T, Martinelli E, et al. Immunotherapy of colorectal cancer: Challenges for therapeutic efficacy. Cancer Treat Rev. 2019;76:22–32.
Article CAS PubMed Google Scholar * Becht E, de Reyniès A, Giraldo NA, Pilati C, Buttard B, Lacroix L, et al. Immune and Stromal Classification of Colorectal Cancer Is Associated with
Molecular Subtypes and Relevant for Precision Immunotherapy. Clin Cancer Res. 2016;22:4057–66. Article CAS PubMed Google Scholar * Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras
(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. Article CAS PubMed PubMed Central Google Scholar * Lito P, Solomon M, Li L-S,
Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–8. Article CAS PubMed PubMed Central Google Scholar * Yaeger R,
Weiss J, Pelster MS, Spira AI, Barve M, Ou SI, et al. Adagrasib with or without Cetuximab in Colorectal Cancer with Mutated KRAS G12C. N Engl J Med. 2023;388:44–54. Article CAS PubMed
Google Scholar * van den Berg I, Smid M, Coebergh van den Braak RRJ, van de Wiel MA, van Deurzen CHM, de Weerd V, et al. A panel of DNA methylation markers for the classification of
consensus molecular subtypes 2 and 3 in patients with colorectal cancer. Mol Oncol. 2021;15:3348–62. Article PubMed PubMed Central Google Scholar * Eide PW, Bruun J, Lothe RA, Sveen A.
CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017;7:16618. Article PubMed PubMed Central Google Scholar * Sawayama H,
Miyamoto Y, Ogawa K, Yoshida N, Baba H. Investigation of colorectal cancer in accordance with consensus molecular subtype classification. Ann Gastroenterol Surg. 2020;4:528–39. Article
PubMed PubMed Central Google Scholar * Fontana E, Eason K, Cervantes A, Salazar R, Sadanandam A. Context matters-consensus molecular subtypes of colorectal cancer as biomarkers for
clinical trials. Ann Oncol. 2019;30:520–7. Article CAS PubMed PubMed Central Google Scholar * Eide PW, Moosavi SH, Eilertsen IA, Brunsell TH, Langerud J, Berg KCG, et al. Metastatic
heterogeneity of the consensus molecular subtypes of colorectal cancer. NPJ Genom Med. 2021;6:59. Article CAS PubMed PubMed Central Google Scholar * Chowdhury S, Hofree M, Lin K, Maru
D, Kopetz S, Shen JP. Implications of Intratumor Heterogeneity on Consensus Molecular Subtype (CMS) in Colorectal Cancer. Cancers. 2021;13:4923. Article CAS PubMed PubMed Central Google
Scholar Download references FUNDING This work was supported by the National Cancer Institute (K22 CA234406 to JPS, and the Cancer Center Support Grant (P30 CA016672), the Cancer Prevention
& Research Institute of Texas (RR180035 to JPS, JPS is a CPRIT Scholar in Cancer Research), and the Col. Daniel Connelly Memorial Fund. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * The
University of Texas MD Anderson Cancer Center, Houston, TX, USA Saikat Chowdhury, Scott Kopetz & John Paul Shen * Caris Life Sciences, Phoenix, AZ, USA Joanne Xiu, Jennifer R. Ribeiro,
Theodore Nicolaides, Jian Zhang & Kelsey A. Poorman * University of California San Francisco Helen Diller Family Comprehensive Cancer Center, San Francisco, CA, USA W. Michael Korn *
Keck School of Medicine, University of Southern California, Los Angeles, CA, USA Heinz-Josef Lenz * Ruesch Center for the Cure of Gastrointestinal Cancers, Lombardi Comprehensive Cancer
Center, Georgetown University, Washington, DC, USA John L. Marshall * Caris Life Sciences, Irving, TX, USA Matthew J. Oberley, George W. Sledge Jr. & David Spetzler Authors * Saikat
Chowdhury View author publications You can also search for this author inPubMed Google Scholar * Joanne Xiu View author publications You can also search for this author inPubMed Google
Scholar * Jennifer R. Ribeiro View author publications You can also search for this author inPubMed Google Scholar * Theodore Nicolaides View author publications You can also search for this
author inPubMed Google Scholar * Jian Zhang View author publications You can also search for this author inPubMed Google Scholar * W. Michael Korn View author publications You can also
search for this author inPubMed Google Scholar * Kelsey A. Poorman View author publications You can also search for this author inPubMed Google Scholar * Heinz-Josef Lenz View author
publications You can also search for this author inPubMed Google Scholar * John L. Marshall View author publications You can also search for this author inPubMed Google Scholar * Matthew J.
Oberley View author publications You can also search for this author inPubMed Google Scholar * George W. Sledge Jr. View author publications You can also search for this author inPubMed
Google Scholar * David Spetzler View author publications You can also search for this author inPubMed Google Scholar * Scott Kopetz View author publications You can also search for this
author inPubMed Google Scholar * John Paul Shen View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS SC: Conceptualization, Investigation,
Methodology, Writing-review and editing. JX: Conceptualization, Data curation, Formal analysis, Investigation, Writing-review and editing. JRR: Investigation, Visualization, Writing-original
draft. TN, MJO, GWS, DS: Resources, Investigation. JZ: Formal analysis, Methodology. WMK: Resources, Investigation. KAP: Validation, Methodology. HL, JLM: Investigation. SK,
Conceptualization, Resources, Supervision, Investigation, Methodology. JPS: Conceptualization, Resources, Supervision, Funding acquisition, Investigation, Methodology, Writing-review and
editing. CORRESPONDING AUTHOR Correspondence to John Paul Shen. ETHICS DECLARATIONS COMPETING INTERESTS HL: BMS, Merck, Bayer, Oncocyte, Fulgent, 3T Bioscience, Invitae, Abalos, AffiniT,
Adagene, Replimune. JM: RESEARCH SUPPORT: 2cureX, OnDose, Arcus Biosciences; PAYMENT/HONORARIA: AstraZeneca, Merck, Bayer, Seagen, Pfizer, Takeda, Taiho Pharmaceutical; CONSULTING OR
ADVISORY ROLE: Caris Life Sciences; OTHER: Indivumed (CMO). GWS: MEETING SUPPORT: Caris Life Sciences; STOCK/STOCK OPTIONS: Syndax, Caris Life Sciences, Tessa Pharm. SK: RESEARCH SUPPORT:
Sanofi, Biocartis, Guardant Health, Array BioPharma, Genentech/Roche, EMD Serono, MedImmune, Novartis, Amgen, Lilly, Daiichi Sankyol CONSULTNG OR ADVISORY ROLE: Genentech, EMD Serono, Merck,
Holy Stone Healthcare, Novartis, Lilly, Boehringer Ingelheim, AstraZeneca/MedImmune, Bayer Health, Redx Pharma, Ipsen, HalioDx, Lutris, Jacobio, Pfizer, Repare Therapeutics, Inivata,
GlaxoSmithKline, Jazz Pharmaceuticals, Iylon, Xilis, Abbvie, Amal Therapeutics, Gilead Sciences, Mirati Therapeutics, Flame Biosciences, Servier, Carina Biotech, Bicara Therapeutics,
Endeavor BioMedicines, Numab, Johnson & Johnson/Janssen, Genomic Health, Frontier Medicines, Replimune, Taiho Pharmaceutical, Cardiff Oncology, Ono Pharmaceutical, Bristol-Myers
Squibb-Medarex, Amgen, Tempus, Foundation Medicine, Harbinger Oncology, Inc, Takeda, CureTeq, Zentalis, Black Stone Therapeutics, NeoGenomics Laboratories, Accademia Nazionale Di Medicina,
Tachyon Therapeutics; STOCK/STOCK OPTIONS: Frontier Medicines; Iylon; Lutris; Navire; Xilis. JPS: RESEARCH SUPPORT: BostonGene, Celsius Therapeutics; CONSULTING OR ADVISORY ROLE: Engine
Biosciences, NaDeNo Nanoscience. JX, JRR, TN, JZ, KAP, MJO, GWS, and DS: employees of Caris Life Sciences. ETHICS APPROVAL AND CONSENT TO PARTICIPATE This study was conducted in accordance
with guidelines of the Declaration of Helsinki, Belmont report, and U.S. Common rule. In keeping with 45 CFR 46.101(b) (4), this study was performed utilizing retrospective, deidentified
clinical data. As such, it is considered Institutional Review Board (IRB) exempt and no patient consent was required. Exempt status was determined by Western IRB. ADDITIONAL INFORMATION
PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
MATERIALS_MERGED SUPPLEMENTARY TABLE S2 SUPPLEMENTARY TABLE S3 SUPPLEMENTARY TABLE S4 RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive
rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed
by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Chowdhury, S., Xiu, J., Ribeiro, J.R. _et al._ Consensus molecular
subtyping of metastatic colorectal cancer expands biomarker-directed therapeutic benefit for patients with CMS1 and CMS2 tumors. _Br J Cancer_ 131, 1328–1339 (2024).
https://doi.org/10.1038/s41416-024-02826-0 Download citation * Received: 29 November 2023 * Revised: 08 August 2024 * Accepted: 12 August 2024 * Published: 04 September 2024 * Issue Date: 02
November 2024 * DOI: https://doi.org/10.1038/s41416-024-02826-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
