A novel lncrna hitt forms a regulatory loop with hif-1α to modulate angiogenesis and tumor growth
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Increasing evidence has indicated that long noncoding RNAs (lncRNAs) play important roles in human diseases, including cancer; however, only a few of them have been experimentally
validated and functionally annotated. Here, we identify a novel lncRNA that we term _HITT_ (HIF-1α inhibitor at translation level). _HITT_ is commonly decreased in multiple human cancers.
Decreased _HITT_ is associated with advanced stages of colon cancer. Restoration of the expression of _HITT_ in cancer cells inhibits angiogenesis and tumor growth in vivo in an
HIF-1α-dependent manner. Further study reveals that _HITT_ inhibits HIF-1α expression, mainly by interfering with its translation. Mechanically, _HITT_ titrates away YB-1 from the 5′-UTR of
_HIF-1α_ mRNA via a high-stringency YB-1-binding motif. The reverse correlation between _HITT_ and HIF-1α expression is further validated in human colon cancer tissues. Moreover, _HITT_ is
one of the most altered lncRNAs upon the hypoxic switch and _HITT_ downregulation is required for hypoxia-induced HIF-1α expression. We further demonstrate that _HITT_ and HIF-1α form an
autoregulatory feedback loop where HIF-1α destabilizes _HITT_ by inducing MiR-205, which directly targets _HITT_ for degradation. Together, these results expand our understanding of the
cancer-associated functions of lncRNAs, highlighting the _HITT_–HIF-1α axis as constituting an additional layer of regulation of angiogenesis and tumor growth, with potential implications
for therapeutic targeting. You have full access to this article via your institution. Download PDF SIMILAR CONTENT BEING VIEWED BY OTHERS LONG NON-CODING RNA PAARH PROMOTES HEPATOCELLULAR
CARCINOMA PROGRESSION AND ANGIOGENESIS VIA UPREGULATING HOTTIP AND ACTIVATING HIF-1Α/VEGF SIGNALING Article Open access 02 February 2022 NF-ΚB-ACTIVATED SPRY4-IT1 PROMOTES CANCER CELL
METASTASIS BY DOWNREGULATING _TCEB1_ MRNA VIA STAUFEN1-MEDIATED MRNA DECAY Article Open access 23 June 2021 LINC01343 TARGETS MIR-526B-5P TO FACILITATE THE DEVELOPMENT OF HEPATOCELLULAR
CARCINOMA BY UPREGULATING ROBO1 Article Open access 12 October 2023 INTRODUCTION Long noncoding RNAs (lncRNAs), defined as transcribed RNA molecules that are >200 nt in length, have
attracted increasing attention [1, 2]. It has been shown that the central functions of lncRNAs encompass every level of the gene expression program involved in regulating multiple biological
processes [3, 4], and aberrant lncRNA expression is associated with complex human diseases, such as cancer [5,6,7]. Further study of lncRNA functions is expected to revise our understanding
of cancer biology and open up new opportunities for cancer treatment [8, 9]. However, the functions and regulation of the vast majority of lncRNAs currently remain completely unknown.
Hypoxia is a hallmark of the solid tumor microenvironment. Activation of the transcription factor hypoxia-inducible factor (HIF-1) enables cancer cells to adapt to the hypoxic environment
via the transactivation of downstream target genes [10, 11]. Vascular endothelial growth factor (VEGF) is a well-established HIF-1 target that plays essential roles in promoting tumor
angiogenesis [12, 13]. Once expressed upon HIF-1α activation, it stimulates the development of new blood vessels within tumors, providing nutrients and oxygen to facilitate tumor growth and
metabolism, clearing metabolic waste and CO2 [13], and also providing a main route for tumor cells to metastasize to distant organs [14]. Increased expression of HIF-1 correlates with the
promotion of angiogenesis, tumor metastasis, and poor prognosis in multiple types of solid tumors, including those of the colon and cervix [15, 16]. Thus, increasing our understanding of the
regulatory mechanisms underlying HIF-1α expression and activation may lead to the development of new anticancer therapies that target HIF-1α [17, 18]. It is well established that HIF-1α is
subjected to proteasome degradation under normal oxygen tension. Under hypoxia, HIF-1α escapes degradation and translocates to the nucleus, where it binds with the hypoxia response element
(HRE) on promoters to initiate gene expression [19]. Although it is believed that such an increase in HIF-1α activity is mainly due to increased protein stability, recent evidence indicates
that translational regulation of HIF-1α after the hypoxic switch may also play an important role. Galban et al. and Hui et al. have both shown that HIF-1α translation accounts for 40–50% of
HIF-1α induction under hypoxia [20, 21]. It has been also reported that the activation of translational signals is correlated with elevated rates of HIF-1α translation and subsequent protein
output [22,23,24]. However, although the regulation of HIF-1α degradation is well understood, much less is known about the regulation of HIF-1α biosynthesis. Interestingly, evidence for the
roles of lncRNAs in regulating HIF-1α activity is beginning to emerge. For example, an _HIF-1α_ lncRNA can directly regulate the stability of _HIF-1α_ mRNA and thus confers poor prognosis
in lung cancer [25]. LncRNAs, such as LincRNA-p21 [26] and RERT [27], regulate HIF-1α indirectly by interfering with the proteasome degradation machinery that regulates HIF-1α. Without
doubt, lncRNAs have added an additional layer of complexity to the HIF-1α-signaling network [28]; however, whether or not HIF-1α biosynthesis can be directly modulated by lncRNAs is still
beyond our understanding. In particular, whether lncRNAs play any roles in HIF-1α’s regulation at the translational level remains unknown. Chromosomal abnormalities are defining features of
solid tumors. Frequently lost chromosome regions in cancer cells can unmask the locations of putative tumor suppressor genes. Notably, loss of 14q32 has been associated with the early onset
and metastatic recurrence of colon cancer, as well as many other types of cancer [29,30,31], suggesting that important tumor suppressor genes might exist in this region. Among them, a
previously uncharacterized lncRNA called linc00637 (we name this gene _HITT_: HIF-1α inhibitor at translation level) has attracted our attention because analysis of The Cancer Genome Atlas
(TCGA) data sets reveals that this gene is significantly downregulated in multiple cancer types. We speculate that _HITT_ may be one such 14q32 entity that possesses potential tumor
suppressive activity and then selected it for further functional analysis. MATERIALS AND METHODS CELL LINES AND TREATMENTS The human colorectal cancer (HCT116, HCT15, SW480, SW620, Colo205,
and HT29), cervical cancer (HeLa, CASKi, SiHa, and C33A) cells, and HUVECs were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) or Dulbecco’s modified Eagle’s medium supplemented
with 10% (v/v) fetal bovine serum (Biological industries). All cells were grown at 37 °C in the humidified incubator with 5% CO2. For hypoxia treatment, cells were cultured in an atmosphere
of 1% O2 at 37 °C in a hypoxia incubator for the indicated periods of time. All cell lines were authenticated and characterized by the supplier and were monitored regularly for their
authenticity (Genetic Testing Biotechnology Corporation, Suzhou, China) and to be free of mycoplasma contamination. TISSUE SAMPLES Human colorectal cancer tissues and their corresponding
adjacent normal controls were collected from the Third Affiliated Hospital of Harbin Medical University in China. Written informed consent was obtained from all patients. The study has been
approved by the Research Ethics Committee of Harbin Medical University, China. Specimens were collected and stored in liquid nitrogen immediately after surgery. XENOGRAFTED TUMOR MODEL IN
VIVO _HITT_ expression was stably restored in HCT116 cells, by using empty vector as a control. The female nude mice between 4 and 5 weeks were purchased from Beijing HFK Bioscience Co.,
Ltd. The cells were paired and 1 × 107 paired cells were inoculated subcutaneously into either side of flank of the same female nude mouse. The tumor volumes were measured every week and
calculated as length × width2 × 0.5. After 4 weeks, the mice were anesthetized and culled. The tumor was carefully removed, photographed, and weighed. All animal procedures were performed
according to protocols approved by the Rules for Animal Experiments published by the Chinese Government (Beijing, China) and approved by the Research Ethics Committee of Harbin Medical
University, China. SMALL INTERFERING RNA (SIRNA) TRANSFECTION siRNA specifically targeting _HITT, YB-1, HIF-1α, HIF-2α,_ or _Ago2_, and nonspecific si-scramble control were synthesized by
GenePharma (Shanghai, China). All transfection experiments were conducted by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The siRNA
sequences are listed in Supplementary Table S1. RNA EXTRACTION AND QRT-PCR Total RNA was isolated using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacture’s protocol.
qRT-PCR was performed in triplicate with an Applied Biosystems Prism 7500 Fast Sequence Detection System using TaqMan universal PCR master mix according to the manufacture’s protocol
(Applied Biosystems Inc, Foster City, CA, USA). Gene expression levels relative to _GAPDH_, _U6_, or _18S_ rRNA were calculated by 2−ΔΔCT method. The primer sequences used in qRT-PCR are
listed in Supplementary Table S2. WESTERN BLOT Total proteins from cell lines or tissue samples were isolated with UREA buffer (8 M Urea, 1 M Thiourea, 0.5% CHAPS, 50 Mm DTT, and 24 Mm
Spermine). Equal amount of proteins were separated at sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After incubation with the indicated primary and secondary
antibodies, signals were visualized by ECL. Membrane was then ready for scanning by Image studio system. Protein quantification was conducted by imageJ software. The antibodies were listed
in Supplementary Table S3. IMMUNOHISTOCHEMISTRY The tissue sample was subjected to antigen retrieval by boiling in 0.01 mol/L citrate buffer for 5 min (min). Slides were then incubated with
anti-CD31 or anti-Ki-67 antibody at room temperature for 1 h. Detection was carried out by the REAL EnVision detection system (Dako) with diaminobenzidine peroxidase serving as chromogen.
After that, the slides were briefly counterstained with hematoxylin before mounting. The images were captured under the same conditions and quantitative analysis using ImageJ software. TUBE
FORMATION ASSAY Each well of 24-well plates was coated with 200 μl of Matrigel (Corning, NY, USA) and polymerized for 1 h at 37 °C. HUVECs and 0.5 ml culture medium were seeded onto the
solidified gel. After 8 h, tube formation was observed and captured with microscopy. LUCIFERASE ASSAY 3× HRE sequence (GCATACGTGGGCGCATACGTGGGCGCATACGTGGGC) was cloned into PGL3-basic
luciferase reporter plasmid, namely HRE-luciferase reporter. HIF-1α wild type (WT) 5′-UTR(1-256), 5′-UTR(1-138, mutant type, MT1), and 5′-UTR(139-256, MT2) were cloned into a PGL3 vector,
respectively, with CMV promoter at upstream of the insert to generate reporter plasmids, namely HIF-1α-5′-UTR (1-256), HIF-1α-5′-UTR (1-138), and HIF-1α-5′-UTR (139-256), respectively. WT or
MiR-205 binding site mutant (MT) _HITT_ were cloned into PMIR-Reporter downstream of luciferase, namely WT _HITT_ reporter and MT _HITT_ reporter, respectively. After the indicated
treatment, cells were harvested and subjected to an assay by using the Dual Luciferase Reporter Assay system (Promega, Madison, USA). The relative luciferase activities were normalized with
the _Renilla_ luciferase activities. CHROMATIN IMMUNOPRECIPITATION (CHIP) Chromatin immunoprecipitation (ChIP) assay was performed to estimate protein binding at specific promoter as
described [32]. Briefly, 1 × 107 cells were collected in lysis buffer A and B (buffer A: 5 mM PIPES, 85 mM KCl, 0.5% NP40. buffer B: 1% SDS, 10 mM EDTA, 50 mM Tris-HCl.) supplemented with
Protease Inhibitor Cocktail (MedChemExpress, Shanghai, China) and sonicated to generate chromatin samples with average fragment sizes of 100–500 bp. Cell lysates were treated with the
indicated antibodies or IgG control at 4 °C overnight. Then, the supernatants were mixed with the blocked Protein A/D sepharose beads to collect the antibody–chromatin complexes. After
washing four times, the immunoprecipitated DNA was eluted and purified for the subsequent qPCR analysis. UV-CROSS-LINKING RNA-IP (CLIP) RNP assay was performed to estimate physiological
interaction between protein and RNA as described [32]. Briefly, 1 × 107 cells were washed and UV cross-linked at 400 mJ/cm2 and collected in lysis buffer supplemented with Protease Inhibitor
Cocktail and RNase inhibitor (Thermo Fisher, Rockford, IL). After the cells were precleared with protein A/G sepharose beads, cell lysates were treated with indicated antibody or IgG
control at 4 °C overnight. Then, the antibody–RNA complexes were collected by using the blocked Protein A/G sepharose beads. After four times washes in the presence of Protease Inhibitor
Cocktail and RNase inhibitor, the immunoprecipitated RNA was eluted and Isolated for the subsequent reaction to reverse transcribed into first-strand cDNA and using for further qRT-PCR
analysis for the target RNA of interest. L-AZIDOHOMOALANINE LABELING TO IDENTIFY NEWLY SYNTHESIZED PROTEINS HeLa cells were washed and incubated for 30 min in methionine‐free medium to
deplete intracellular methionine, followed by incubation with 50 μM l-azidohomoalanine (AHA, Invitrogen, Carlsbad, CA, USA) for 4 h under hypoxia. Cells were washed twice with PBS, lysis
buffer were added, and then sonicated for two cycles of 30 s each and centrifuged cell at 13,000 _g_. Fifty micrograms of lysates were subjected to Click reactions according to the
manufacturer’s instructions (Click-iT® Protein Reaction Buffer Kit; Invitrogen, Carlsbad, CA, USA). Total proteins from Click reactions were precipitated with methanol/chloroform and
resolubilized in 50 mM Tris, pH 7.5, 0.01% SDS, and then incubated with 50 μl of Streptavidin coupled magnetic beads for 5 h at room temperature. Streptavidin coupled magnetic beads were
incubated in 50 μl of 2× loading blue for 10 min at 100 °C and detected by western blots. IN VITRO RNA PULL-DOWN ASSAY Biotin-labeled RNA were in vitro synthesized by Biotin RNA Labeling Mix
(Roche, St Louis, MO, USA). After treatment with RNase-free DNase I, Biotin-labeled RNA was heated at 95 °C for 2 min and then cooled on ice for 3 min. Then secondary structure recovered
Biotin-labeled RNA reacted with streptavidin agarose beads (Invitrogen, Carlsbad, CA, USA) overnight. The fresh HCT116 and HeLa lysates were collected and incubated with RNA-captured beads
at 4 °C for 1 h. Beads were briefly washed for five times with wishing buffer (50 mM Tris-HCl (PH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.05% NP40). The resulting beads were boiled at 95 °C for 5
min in SDS loading buffer and then subjected to western blot analysis. Than the samples were detected on 15% polyacrylamide TBE gel at 4 °C for 60 min at 200 V. EMSA In vitro synthesized RNA
oligonucleotides were heated at 70 °C for 5 min and then cooled on ice for 2 min ready for EMSA. One picomole in vitro synthesized dsDNA and 20 pmol prepared ssRNA were incubated in 10 μl
buffer that contain 1 μl 10× triplex forming buffer (100 mM Tris pH 7.5, 25 mM NaCl, and MgCl2) at 37 °C for 30 min. In control assay, triplex reaction was treated with either 5 U of RNase H
or 20 mg/ml RNase A at 37 °C for 30 min. POLYSOME FRACTIONATION Cells were treated with 100 μg/ml cycloheximide (CHX) for 5 min and then lysed in polysome lysis buffer (15 mM Tris-HCL PH
7.5, 15 mM MgCl2, 0.3 M NaCl, 1% Triton X-100, 0.1 U/μl RNA inhibitor, 100 μg/ml CHX, 1 μg/ml Heparin, and 1× protease inhibitor cocktail). The sucrose gradient samples were obtained by
centrifuge at 39,000 rpm for 2 h at 4 °C using SW40Ti rotor in a Beckman Coulter and monitored by using ultraviolet spectrophotometer at 254 nm. RNA in each sucrose gradient was collected
and extracted in 3 volumes of Trizol, followed by qRT-PCR assay for the indicated genes. ELISA VEGF concentration of cell culture medium was measured by ELISA using the Quantikine human VEGF
ELISA Kit according to the manufacturer’s instructions (R&D Systems, Minneapolis, MN, USA). The sample absorbance was determined at 450 and 540 nm. Each experiment was repeated three
times. STATISTICAL ANALYSIS Statistical analysis was done by GraphPad software, version 5. Data are presented as the means ± standard error of the means (SEM) or standard Deviation (SD).
Student _t_ test was applied to assess the statistical significance. Correlations were calculated according to Spearmen or Pearson correlation. _p_ value < 0.05 were considered
significant. RESULTS LNCRNA _HITT_ IS COMMONLY REDUCED IN HUMAN CANCERS Rapid amplification of 3′- and 5′-complementary DNA ends (3′- and 5′-RACE) assays confirmed that _HITT_ (linc00637,
Fig. S1A) is a 2050-nt transcript possessing a polyadenylation site (Fig. S1B), with the expected size (~2 kb) as revealed by northern blot assay (Fig. S1C). _HITT_ received a coding
potential value of 0.387, similar to other well-characterized lncRNAs and much lower than protein-coding genes, as analyzed by Coding Potential Calculation (Fig. S1D). Comparable amounts of
_HITT_ were detected in both the nucleus and cytoplasm, using cytoplasmic _GAPDH_ mRNA and the nuclear lncRNA _MALAT1_ [33] as controls (Fig. S1E). Remarkably, qRT-PCR results revealed that
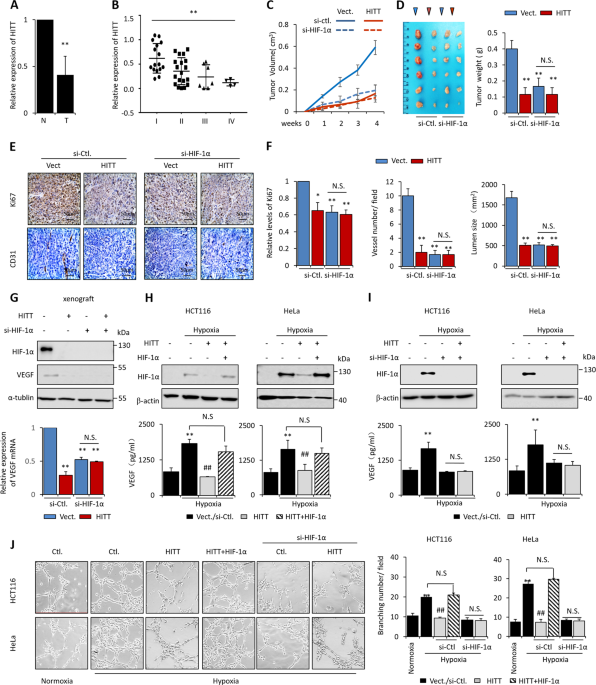
_HITT_ was significantly downregulated in the colon cancer tissues analyzed relative to the paired normal controls (Fig. 1a). Absolute values of fold-change increased according to TNM stages
(Fig. 1b). _HITT_ levels were much lower in colon cancer cell lines than in pooled independent normal tissue controls (Fig. S2A). Furthermore, _HITT_ levels were reduced in the cervical
(_n_ = 6) and renal cell carcinoma tissues (_n_ = 17), and cervical cancer cell lines examined (Fig. S2B–C). Analysis of the TCGA database revealed that _HITT_ is downregulated in multiple
cancer types, such as colon cancer, thyroid cancer, and chromophobe renal cell carcinoma (Fig. S2D), and that decreased _HITT_ is associated with advanced stages of bladder, breast, and
liver cancers (Fig. S2E). Lower levels of _HITT_ also predict poor clinical outcomes in lung and rectal cancers (Fig. S2F). These findings collectively indicate that _HITT_ is commonly
downregulated in human cancers. _HITT_ SUPPRESSES ANGIOGENESIS AND TUMOR GROWTH IN A HIF-1Α-DEPENDENT MANNER To understand its functional significance, single clones of HeLa and HCT116 cells
stably expressing _HITT_ at levels similar to those of normal controls were used for further analysis (Fig. S3A). However, no obvious differences in the anchorage-dependent or -independent
growth capabilities was observed after restoration of _HITT_ expression (Fig. S3B–C). Whereas _HITT_ expression significantly reduced the tumor volumes and weights of HCT116 xenografts
implanted in nude mice (Fig. 1c, d). These data suggest that _HITT_ inhibits tumor growth, possibly by antagonizing detrimental microenvironmental factors. Hypoxia is a principle
microenvironment factor that initiates angiogenesis and promotes tumor growth. Given that HIF-1α is a master regulator of tumor angiogenesis, we examined whether the antitumor effect of
_HITT_ relies on the presence of HIF-1α protein. To this end, HIF-1α expression was inhibited using siRNA specifically targeted to HIF-1α. In line with previous reports, HIF-1α knockdown
(KD) abolished HIF-1α expression and significantly reduced the tumor growth of HCT116 xenografts (2.5–3-fold, Fig. 1c, d). Importantly, the effect of _HITT_ on tumor growth was completely
abrogated by HIF-1α KD (Fig. 1c, d). Consistently, Ki-67 intensity and vascular density were reduced with _HITT_ expression or HIF-1α KD, and no further reduction was observed upon their
combination (Fig. 1e, f). _VEGF_, a known HIF-1α downstream pro-angiogenesis target [13], was inhibited by _HITT_ (Fig. 1g), paralleling the decreased tumor vasculature (Fig. 1e, f) and
HIF-1α protein levels (Figs. 1g and S3D). The effect of _HITT_ on HIF-1α-induced angiogenesis was verified using in vitro models. The conditioned medium from cells cultured under hypoxia
significantly stimulated the branching abilities of HUVECs relative to the normoxia control (Fig. 1h–j). _HITT_ transfection abrogated conditioned medium-induced HUVEC branching (Fig. 1j).
HIF-1α KD also abrogated hypoxia-induced VEGF expression and HUVEC branching, and, interestingly, completely abolished the antiangiogenic effect of _HITT_ (Fig. 1i). In contrast, HIF-1α
overexpression rescued _HITT_-repressed HUVECs branching (Figs. 1h, j). Collectively, the findings indicate that _HITT_ inhibits angiogenesis and tumor growth by interfering with
HIF-1α-dependent signals. _HITT_ INHIBITS HIF-1Α EXPRESSION AND ACTIVITY We next sought out to understand how _HITT_ regulates HIF-1α signals. The results show that _HITT_ markedly
diminished hypoxia-induced HIF-1α expression by approximately five to sixfold (Fig. 2a, b), while _HITT_ KD increased HIF-1α under normoxia (Fig. 2c–e). We also noticed that _HITT_
consistently inhibited _HIF-1α_ mRNA expression by approximately twofold (lower panel, Fig. 2b), while _HITT_ KD promoted it (lower panel, Fig. 2e). However, the effect of _HITT_’s
regulation of HIF-1α protein was much more dramatic than its effect on _HIF-1α_ mRNA under the same conditions (Fig. 2b, e). These data indicate that _HITT_ mainly regulates HIF-1α at
protein levels. The inhibitory effect of _HITT_ on HIF-1α is likely specific, as _HITT_ overexpression or KD failed to change the mRNA or protein expression levels of its family member
HIF-2α (Fig. S4A–B). More importantly, the transcriptional activity of HIF-1α, as evidenced by HRE-dependent reporter activity, was markedly increased by hypoxia and this effect was
abolished in a stable _HITT_-expressing cell line (Fig. 2f), while ectopic HIF-1α expression dampened the effect of _HITT_ (Fig. 2f). In addition, the expression of well-established HIF-1α
targets, such as VEGF and angiogenin-1, were increased by hypoxia (Fig. 2g), which was significantly blunted by _HITT_ (Fig. 2g). These data collectively illustrate that _HITT_ inhibits the
transcriptional activity of HIF-1α, mainly by regulating its protein levels. _HITT_ INHIBITS HIF-1Α TRANSLATION _HITT_ is unlikely to affect HIF-1α protein stability. As shown, HIF-1α
decayed at similar rates after cells were treated with the protein synthesis inhibitor CHX in a _HITT_-transfected stable line and control line (Fig. 3a). _HITT_ similarly inhibited HIF-1α
regardless of MG132 (Fig. 3b). We further compared the efficiency of HIF-1α protein synthesis by polysome fractionation. _HITT_ expression did not change the polysome distribution profile
(Fig. 3c), while polysome-associated _HIF-1α_ mRNA was increased with hypoxia (Fig. 3c), which is in line with a previous report [21]. Importantly, ectopic _HITT_ expression compromised
hypoxia-induced association of _HIF-1α_ mRNA with polysomes (Fig. 3c), while _HITT_ KD increased their association (Fig. 3d). In contrast, the distribution of _GAPDH_ mRNA remained
unaltered, regardless of oxygen concentration or _HITT_ levels (Fig. 3d). The role of _HITT_ in regulating HIF-1α translation was further verified by a Click chemistry and AHA-labeled assay.
As shown, _HITT_ overexpression inhibited the newly synthesized HIF-1α protein (Fig. 3e), while _HITT_ KD increased it under hypoxia (Fig. 3f). Newly synthesized α-tubulin served as a
negative control, and was not changed by ectopic _HITT_ expression or KD (Fig. 3e, f). Therefore, _HITT_ inhibits _HIF-1α_ mRNA translation. _HITT_ ACTS BY SERVING AS A DECOY OF HIF-1Α
TRANSLATIONAL REGULATOR YB-1 Recent evidence has shown that YB-1 promotes HIF-1α translation by directly interacting with its 5′-UTR [34, 35]. Although _HITT_ had no obvious impact on YB-1
protein expression both in vitro and in vivo in HCT116 nude mice xenografts (Fig. 4a and Fig. S5A), eight YB-1-binding sequences were predicted in _HITT_ via a high-stringency search using
an online bioinformatics tool (Fig. 4b). YB-1 KD reduced HIF-1α levels and _HITT_ lost its ability to affect HIF-1α expression in YB-1 KD cells (Fig. 4a). This brought us to ask whether
_HITT_ regulates HIF-1α transcription by competitively binding with YB-1. As shown, YB-1 was co-precipitated with biotin-_HITT_ in vitro (Fig. 4c). Both _HITT_ and the HIF-1α 5′-UTR, but not
_GAPDH_ mRNA, were physically associated with YB-1, as revealed by CLIP assays (Figs. 4d, e and S5B). The enrichment of _HITT_ was significantly decreased, while that of the 5′-UTR was
increased in response to the hypoxic switch (Figs. 4d, e and S5B). Ectopic _HITT_ expression restored the association of _HITT_ with YB-1 under hypoxia and simultaneously abrogated the
hypoxia-promoted association of the HIF-1α 5′-UTR with YB-1 (Figs. 4d, e and S5B). In contrast, _HITT_ KD led to promotion of the association between YB-1 and the HIF-1α 5′-UTR (Fig. 4f).
_HITT_ containing the predicted YB-1-binding motif contributed to its associated with YB-1 (Fig. 4g). A _HITT_ mutant that had lost the potential YB-1 binding sites completely lost its
regulatory effect towards HIF-1α (Figs. 4h and S5C). These data together suggest that binding of YB-1 is required for _HITT_-mediated HIF-1α inhibition. Furthermore, both YB-1 KD and ectopic
_HITT_ expression prevented hypoxia-induced WT HIF-1α-5′-UTR-luciferase reporter activity, while no further reduction was detected when they were introduced in combination (Figs. 4i and
S5D). The truncated mutant reporters MT1 and MT2, were also induced by hypoxia but to a much lower extent (Figs. 4i and S5D). However, only MT2 responded to YB-1 KD and ectopic _HITT_
expression, suggesting that sequences 2 and 3 in MT2 are the major YB-1 binding sites in HIF-1α-5′-UTR (Figs. 4i and S5D). Altogether, _HITT_ directly binds with YB-1 and prevents its
association with HIF-1α 5′-UTR, leading to reduced HIF-1α translation. HIF-1Α LOWERS _HITT_’S STABILITY VIA A FEEDBACK LOOP To understand the physiological relevance of _HITT_ in regulating
HIF-1α under hypoxia, we examined the change in _HITT_ expression under hypoxia. Interestingly, _HITT_ was significantly reduced under hypoxia and the extent of the reduction was similar to
that of the most dramatically altered lncRNAs reported previously [28] (Fig. 5a). In addition, _HITT_ reduction was oxygen dose- and hypoxia duration-dependent (Fig. 5b, c). si-HIF-1α
reduced HIF-1α levels and simultaneously restored the expression of _HITT_ (Figs. 5d and S6A). In contrast, ectopic HIF-1α expression lowered _HITT_ (Figs. 5e and S6B). HIF-2α did not show
any obvious impact on the expression of _HITT_ (Fig. S6C–D), suggesting that _HITT_ is specifically regulated by HIF-1α via a feedback loop. Hypoxia/HIF-1α-induced _HITT_ suppression is
unlikely to occur at the transcriptional level. Firstly, no HRE motif was identified in _HITT_’s promoter. Secondly, neither ectopic HIF-1α expression nor KD had a significant effect on
_HITT_ promoter-driven luciferase activity (Figs. 5d, e and S6A–B). Nonetheless, the half-life of _HITT_, but not _GAPDH_ mRNA, was decreased by hypoxia. HIF-1α KD blocked the effect of
hypoxia (Figs. 5f and S6E). Consistently, comparable _HITT_ levels were detected under normoxia and hypoxia in HIF-1α-deficient 786-O cells (Fig. S6F–G). These data demonstrate that HIF-1α
governs _HITT_’s stability. HIF-1Α PROMOTES _HITT_’S DECAY BY MIR-205-MEDIATED RNA DEGRADATION We next sought to investigate the mechanisms underlying HIF-1α-mediated _HITT_ destabilization.
Two independent bioinformatics tools predicted that _HITT_ is a candidate target of a number of microRNAs, of which MiR-205 was the most likely (Fig. 6a). In addition, among the microRNAs
that indeed possess the ability to inhibit _HITT_ expression as validated by qRT-PCR (Fig. S7A), only MiR-205 was induced over twofold by hypoxia (Fig. S7B). We also found that the
expression of MiR-205 was dependent on the levels of HIF-1α, because hypoxia-induced MiR-205 was completely abolished by siRNA-mediated HIF-1α KD in both HCT116 and HeLa cells (Fig. 6b).
Thus, it is possible that MiR-205 mediates HIF-1α-induced downstream events, such as _HITT_ destabilization. Therefore, we further investigated the impacts of MiR-205 on _HITT_ expression.
As shown, a MiR-205 inhibitor prevented hypoxia-induced MiR-205 and also restored reduced levels of _HITT_ under hypoxia (Figs. 6c and S7C), whereas the induction of MiR-205 under normoxia
by transfecting MiR-205 mimics significantly reduced _HITT_ (Fig. 6d). MiR-205 mimics also abolished HIF-1α KD-induced _HITT_ recovery under hypoxia (Figs. 6e and S7D). Furthermore, the
activity of WT _HITT_ luciferase reporter was decreased by hypoxia. MiR-205 inhibitor completely reversed the inhibitory effect of hypoxia on WT _HITT_ luciferase activity (Figs. 6g and
S7E). However, neither hypoxia nor the MiR-205 inhibitor influenced the luciferase activity of the MiR-205-binding defective MT reporter (Figs. 6g and S7E). In contrast, MiR-205
significantly reduced WT, but not MT, _HITT_ luciferase activity (Figs. 6h and S7F). Furthermore, the direct association between MiR-205 and _HITT_ was verified by pull-down biotinylated
precursor MiR-205 followed by qRT-PCR analysis of _HITT_ (Fig. 6i). Moreover, KD of Ago2, an important component of the RNA-induced silencing complex/MiR-205, produced a similar effect on
_HITT_ as MiR-205 inhibitors (Figs. 6j and S7G). RIP Ago2 showed that _HITT_ was associated with Ago2 (Fig. 6k). Although the expression levels of Ago2 were not changed under hypoxia (Fig.
S7H), levels of Ago2-associated _HITT_ were increased (Fig. 6k), under which conditions _HITT_ was destabilized. A MiR-205 inhibitor reduced the hypoxia-induced association between Ago2 and
_HITT_ (Fig. 6k), with MiR-205 losing its ability to affect _HITT_ expression in Ago2 KD cells (Fig. 6l). Consistently, the MiR-205 inhibitor or Ago2 KD prolonged _HITT_’s half-life, while
no further change was observed when they were combined (Fig. 6m). Collectively, these data suggest a model where Ago2/miR-205 physiologically binds with _HITT_, facilitating MiR-205-mediated
_HITT_ destabilization. _HITT_ DOWNREGULATION IS CORRELATED WITH HIGH LEVELS OF HIF-1Α IN HUMAN COLON CANCER TISSUES Having uncovered a novel _HITT_–HIF-1α axis, we verified the correlation
between HIF-1α and _HITT_ expression in human cancer tissues. The protein levels of HIF-1α and its target, _VEGF_, were examined in human colon cancer tissues and matched adjacent controls
(_n_ = 46). HIF-1α and VEGF protein levels were commonly increased in colon cancer tissues compared with their matched normal controls (Fig. 7a–c). Interestingly, reduced _HITT_ levels were
significantly correlated with increased HIF-1α protein and _VEGF_ mRNA expression (Fig. 7d, e). These findings, combined with our in vitro and xenograft data, demonstrate that _HITT_
regulates HIF-1α at the translational level, which results in reduced expression of HIF-1α and its targets. DISCUSSION The discovery of the functional significance of protein-coding genes
has led to great successes in the development of targeted therapies against cancer. Increasing evidence has suggested that nonprotein-coding genes account for the overwhelming majority of
RNA transcripts and can drive important cancer phenotypes. Such discoveries have sparked great interest in the functions and regulation of lncRNAs [9, 36], with a view to improving the
diagnosis and treatment of cancers. _HITT_ is one such as yet uncharacterized lncRNA in the human genome. We found that the expression of _HITT_ was downregulated in multiple human tumors.
Although it had no obvious impact on tumor growth in vitro, _HITT_ elicited a marked suppressive effect in vivo by influencing angiogenesis. The key mechanism that we elucidated as
accounting for this function of _HITT_ is that it inhibits HIF-1α expression (Fig. 7f). The significance of our in vitro and animal experiment data is further strengthened by the fact that a
reverse association between _HITT_ and key components of this pathway, i.e. HIF-1α protein and VEGF transcripts, were detected in human colon cancer tissues. Of note, angiogenesis and
increased HIF-1α levels are associated with poor prognosis in multiple cancers [11, 37]. In line with this notion, _HITT_ levels were inversely correlated with advanced stages of colon
cancers. These data encourage the future development of _HITT_-based cancer therapies for patients with increased HIF-1α, who are at high risk of metastasis [38], an outcome that results in
~90% of cancer-related deaths. HIF-1α is a core transcription factor for the adaptive survival of cancer cells under hypoxia. The mechanisms underlying HIF-1α protein stability have been
studied extensively and are also considered to be critical for the accumulation of HIF-1α under hypoxia [39]. Here, we have demonstrated that _HITT_ is an important HIF-1α negative
regulator. Intriguingly, the action of _HITT_ is mainly fulfilled via its regulating of HIF-1α translation, but not its protein stability (Fig. 7f). This mechanism is distinct from
lncRNA-mediated HIF-1α regulation reported previously, through which lncRNAs mainly effect the HIF-1α protein stabilization machinery [26] or sponging microRNAs target HIF-1α [40]. LncRNAs
can act as ‘molecular decoys for RNA-binding proteins, which are themselves regulatory factors of gene expression [41, 42]. YB-1 is a well-characterized translational regulator. It has
recently been reported to regulate HIF-1α translation. Like the typical YB-1 target SNAIL1 [43], HIF-1α 5′-UTR contains a complex stem-loop structure. Thereby, El-Naggar et al. have proposed
that YB-1 binding may unwind such secondary structures, leading to the promotion of HIF-1α translation [34]. Our study reveals an additional level of regulation for YB-1-mediated HIF-1α
translation via lncRNA. _HITT_ prevents or interrupts the interaction of YB-1 with HIF-1α 5′-UTR to suppress HIF-1α expression. In fact, the lncRNAs CAR10 [44], TP53TG1 [45], and GAS5 [46]
have been recently documented as regulators of YB-1 activity. These lncRNAs act by controlling YB-1’s localization or protein stability. In contrast to these lncRNAs, _HITT_ inhibits YB-1
via a distinct mechanism by acting as a protein decoy. These findings combined suggest that lncRNAs may be important components of the regulatory machinery for modulating YB-1’s activity by
diverse means. It will be also interesting to explore in the future whether _HITT_ is involved in regulating multiple functions of YB-1 [47]. Given its importance in sensing hypoxic tension,
it is not surprising that cells utilize feedback mechanisms to precisely control HIF-1α signals. In fact, a number of hypoxia-responsive lncRNAs contain HREs and subjected to direct
regulation by HIF-1α [48]. However, the feedback regulation of HIF-1α by _HITT_, identified here, is unlikely to occur through a direct mechanism [49]. Our data suggest that MiR-205 is a
primary microRNA to inhibit _HITT_ under hypoxia. However, _HITT_ may be regulated by additional microRNAs, as more microRNAs are predicted to undergo complementary base pairing with _HITT_.
It will be interesting to explore the roles of these microRNAs on _HITT_ expression under different physiological and pathological conditions in future studies. Furthermore, besides
microRNAs, lncRNAs may also be regulated by typical transcription factors [26] or RNA-binding proteins [33]. The precise molecular mechanism of the downregulation of _HITT_ requires further
research. Taken together, we have identified _HITT_, a novel lncRNA. Our data highlight the significance of the newly identified _HITT_–HIF-1α axis in regulating angiogenesis and tumor
growth. Detailed dissection of the molecular mechanisms of _HITT_-mediated HIF-1α synthesis have provided insights into novel strategies to inhibit HIF-1α by restoring _HITT_ activity.
REFERENCES * Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. Article CAS PubMed PubMed Central Google Scholar * Iyer MK, Niknafs YS, Malik R,
Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. Article CAS PubMed PubMed Central Google Scholar * Wang
KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. Article CAS PubMed PubMed Central Google Scholar * Guttman M, Rinn JL. Modular regulatory principles
of large non-coding RNAs. Nature. 2012;482:339–46. Article CAS PubMed PubMed Central Google Scholar * Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61.
Article CAS PubMed Google Scholar * Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. Article CAS PubMed PubMed Central Google Scholar *
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. Article CAS PubMed PubMed Central Google Scholar * Evans JR, Feng FY, Chinnaiyan
AM. The bright side of dark matter: lncRNAs in cancer. J Clin Investig. 2016;126:2775–82. Article PubMed PubMed Central Google Scholar * Adams BD, Parsons C, Walker L, Zhang WC, Slack
FJ. Targeting noncoding RNAs in disease. J Clin Investig. 2017;127:761–71. Article PubMed PubMed Central Google Scholar * Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science.
2016;352:175–80. Article CAS PubMed PubMed Central Google Scholar * Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11:393–410. Article CAS PubMed Google
Scholar * Ma L, Chen ZM, Li XY, Wang XJ, Shou JX, Fu XD. Nucleostemin and ASPP2 expression is correlated with pituitary adenoma proliferation. Oncol Lett. 2013;6:1313–8. Article CAS
PubMed PubMed Central Google Scholar * Jain RK. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell. 2014;26:605–22. Article CAS PubMed
PubMed Central Google Scholar * Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–87. Article CAS PubMed Google Scholar * Birner P,
Schindl M, Obermair A, Plank C, Breitenecker G, Oberhuber G. Overexpression of hypoxia-inducible factor 1alpha is a marker for an unfavorable prognosis in early-stage invasive cervical
cancer. Cancer Res. 2000;60:4693–6. CAS PubMed Google Scholar * Auyeung KK, Ko JK. Angiogenesis and oxidative stress in metastatic tumor progression: pathogenesis and novel therapeutic
approach of colon cancer. Curr Pharm Des. 2017;23:3952–61. Article CAS PubMed Google Scholar * Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for
cancer therapy. Trends Pharm Sci. 2012;33:207–14. Article CAS PubMed Google Scholar * Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. Article CAS PubMed
Google Scholar * Dengler VL, Galbraith M, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49:1–15. Article CAS PubMed Google
Scholar * Galban S, Kuwano Y, Pullmann R Jr, Martindale JL, Kim HH, Lal A, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol.
2008;28:93–107. Article CAS PubMed Google Scholar * Hui AS, Bauer AL, Striet JB, Schnell PO, Czyzyk-Krzeska MF. Calcium signaling stimulates translation of HIF-alpha during hypoxia.
FASEB J. 2006;20:466–75. Article CAS PubMed Google Scholar * Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via
p44/42 MAPK and NF-kappaB. Biochemical J. 2006;396:517–27. Article CAS Google Scholar * Mi C, Ma J, Wang KS, Zuo HX, Wang Z, Li MY, et al. Imperatorin suppresses proliferation and
angiogenesis of human colon cancer cell by targeting HIF-1alpha via the mTOR/p70S6K/4E-BP1 and MAPK pathways. J Ethnopharmacol. 2017;203:27–38. Article CAS PubMed Google Scholar * Zhou
C, Huang C, Wang J, Huang H, Li J, Xie Q, et al. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via
modulating PHLPP1 transcription and HIF-1alpha translation. Oncogene. 2017;36:3878–89. Article CAS PubMed PubMed Central Google Scholar * Singh L, Aldosary S, Saeedan AS, Ansari MN,
Kaithwas G. Prolyl hydroxylase 2: a promising target to inhibit hypoxia-induced cellular metabolism in cancer cells. Drug Discov Today. 2018;23:1873–82. * Yang F, Zhang H, Mei Y, Wu M.
Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol cell. 2014;53:88–100. Article CAS PubMed Google Scholar * Kapinova A, Kubatka P, Zubor P,
Golubnitschaja O, Dankova Z, Uramova S, et al. The hypoxia-responsive long non-coding RNAs may impact on the tumor biology and subsequent management of breast cancer. Biomedicine
Pharmacother. 2018;99:51–8. Article CAS Google Scholar * Choudhry H, Harris AL. Advances in hypoxia-inducible factor biology. Cell Metab. 2018;27:281–98. Article CAS PubMed Google
Scholar * Taniwaki M, Matsuda F, Jauch A, Nishida K, Takashima T, Tagawa S, et al. Detection of 14q32 translocations in B-cell malignancies by in situ hybridization with yeast artificial
chromosome clones containing the human IgH gene locus. Blood. 1994;83:2962–9. Article CAS PubMed Google Scholar * Bando T, Kato Y, Ihara Y, Yamagishi F, Tsukada K, Isobe M. Loss of
heterozygosity of 14q32 in colorectal carcinoma. Cancer Genet Cytogenet. 1999;111:161–5. Article CAS PubMed Google Scholar * Dodd LE, Sengupta S, Chen IH, den Boon JA, Cheng YJ, Westra
W, et al. Genes involved in DNA repair and nitrosamine metabolism and those located on chromosome 14q32 are dysregulated in nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev.
2006;15:2216–25. Article CAS PubMed Google Scholar * Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in
triple-negative breast cancer. Nat Struct Mol Biol. 2016;23:522–30. Article CAS PubMed PubMed Central Google Scholar * Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, et al. Long
non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014;5:e1008. Article CAS PubMed PubMed Central Google Scholar * El-Naggar AM, Veinotte
CJ, Cheng H, Grunewald TG, Negri GL, Somasekharan SP, et al. Translational activation of HIF1alpha by YB-1 promotes sarcoma metastasis. Cancer Cell. 2015;27:682–97. Article CAS PubMed
Google Scholar * Ivanova IG, Park CV, Yemm AI, Kenneth NS. PERK/eIF2alpha signaling inhibits HIF-induced gene expression during the unfolded protein response via YB1-dependent regulation of
HIF1alpha translation. Nucleic Acids Res. 2018;46:3878–90. Article CAS PubMed PubMed Central Google Scholar * Velagapudi SP, Cameron MD, Haga CL, Rosenberg LH, Lafitte M, Duckett DR,
et al. Design of a small molecule against an oncogenic noncoding RNA. PNAS. 2016;113:5898–903. Article CAS PubMed PubMed Central Google Scholar * Baba Y, Nosho K, Shima K, Irahara N,
Chan AT, Meyerhardt JA, et al. HIF1A overexpression is associated with poor prognosis in a cohort of 731 colorectal cancers. Am J Pathol. 2010;176:2292–301. Article CAS PubMed PubMed
Central Google Scholar * Unwith S, Zhao H, Hennah L, Ma D. The potential role of HIF on tumour progression and dissemination. Int J cancer. 2015;136:2491–503. Article CAS PubMed Google
Scholar * Shenoy N, Pagliaro L. Sequential pathogenesis of metastatic VHL mutant clear cell renal cell carcinoma: putting it together with a translational perspective. Ann Oncol.
2016;27:1685–95. Article CAS PubMed Google Scholar * Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci.
2014;127:1585–94. Article CAS PubMed PubMed Central Google Scholar * Zhang J, Zhang Q. VHL and hypoxia signaling: beyond HIF in cancer. Biomedicines. 2018;6:1–13. * Yang J, Harris AL,
Davidoff AM. Hypoxia and hormone-mediated pathways converge at the histone demethylase KDM4B in cancer. Int J Mol Sci. 2018;19:1–11. * Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink
D, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–15.
Article CAS PubMed Google Scholar * Cox TR, Erler JT, Rumney RMH. Established models and new paradigms for hypoxia-driven cancer-associated bone disease. Calcif Tissue Int.
2018;102:163–73. Article CAS PubMed Google Scholar * Salem A, Asselin MC, Reymen B, Jackson A, Lambin P, West CML, et al. Targeting hypoxia to improve non-small cell lung cancer outcome.
J Natl Cancer Inst. 2018;110:14–30. * Jahanban-Esfahlan R, de la Guardia M, Ahmadi D, Yousefi B. Modulating tumor hypoxia by nanomedicine for effective cancer therapy. J Cell Physiol.
2018;233:2019–31. Article CAS PubMed Google Scholar * Lasham A, Print CG, Woolley AG, Dunn SE, Braithwaite AW. YB-1: oncoprotein, prognostic marker and therapeutic target? Biochemical J.
2013;449:11–23. Article CAS Google Scholar * Choudhry H, Harris AL, McIntyre A. The tumour hypoxia induced non-coding transcriptome. Mol Asp Med. 2016;47-48:35–53. Article CAS Google
Scholar * Wang X, Yu M, Zhao K, He M, Ge W, Sun Y, et al. Upregulation of MiR-205 under hypoxia promotes epithelial-mesenchymal transition by targeting ASPP2. Cell Death Dis. 2016;7:e2517.
Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This work was funded by the National Natural Science Foundation of China (grant numbers 31301131,
31741084, and 31871389) and the Basic Science Foundation of Science and Technology Innovation Commission in Shenzhen (grant number JCYJ20170811154452255). AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * School of Life Science and Technology, Harbin Institute of Technology, 150001, Harbin, Heilongjiang Province, China Xingwen Wang, Kunming Zhao, Qingyu Lin, Huayi Li, Xuting
Xue, Wenjie Ge, Hongjuan He, Dong Liu, Qiong Wu & Ying Hu * The Affiliated Tumor Hospital of Harbin Medical University, 150001, Harbin, Heilongjiang Province, China Li Li * State Key
Laboratory of Robotics and Systems, Harbin Institute of Technology, 2 Yikuang, 150001, Harbin, China Hui Xie * Shenzhen Graduate School of Harbin Institute of Technology, 518055, Shenzhen,
China Ying Hu Authors * Xingwen Wang View author publications You can also search for this author inPubMed Google Scholar * Li Li View author publications You can also search for this author
inPubMed Google Scholar * Kunming Zhao View author publications You can also search for this author inPubMed Google Scholar * Qingyu Lin View author publications You can also search for
this author inPubMed Google Scholar * Huayi Li View author publications You can also search for this author inPubMed Google Scholar * Xuting Xue View author publications You can also search
for this author inPubMed Google Scholar * Wenjie Ge View author publications You can also search for this author inPubMed Google Scholar * Hongjuan He View author publications You can also
search for this author inPubMed Google Scholar * Dong Liu View author publications You can also search for this author inPubMed Google Scholar * Hui Xie View author publications You can also
search for this author inPubMed Google Scholar * Qiong Wu View author publications You can also search for this author inPubMed Google Scholar * Ying Hu View author publications You can
also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Ying Hu. ETHICS DECLARATIONS CONFLICT OF INTEREST The authors declare that they have no conflict of
interest. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Edited by I. Amelio
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY FIGURE 1 SUPPLEMENTARY FIGURE 2 SUPPLEMENTARY FIGURE 3 SUPPLEMENTARY FIGURE 4 SUPPLEMENTARY
FIGURE 5 SUPPLEMENTARY FIGURE 6 SUPPLEMENTARY FIGURE 7 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Wang, X., Li, L., Zhao, K. _et al._ A novel LncRNA
_HITT_ forms a regulatory loop with HIF-1α to modulate angiogenesis and tumor growth. _Cell Death Differ_ 27, 1431–1446 (2020). https://doi.org/10.1038/s41418-019-0449-8 Download citation *
Received: 18 February 2019 * Revised: 21 October 2019 * Accepted: 21 October 2019 * Published: 07 November 2019 * Issue Date: April 2020 * DOI: https://doi.org/10.1038/s41418-019-0449-8
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative
