Actin-driven nanotopography promotes stable integrin adhesion formation in developing tissue
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Morphogenesis requires building stable macromolecular structures from highly dynamic proteins. Muscles are anchored by long-lasting integrin adhesions to resist contractile force.
However, the mechanisms governing integrin diffusion, immobilization, and activation within developing tissues remain elusive. Here, we show that actin polymerization-driven membrane
protrusions form nanotopographies that enable strong adhesion at Drosophila muscle attachment sites (MASs). Super-resolution microscopy reveals that integrins assemble adhesive belts around
Arp2/3-dependent actin protrusions, forming invadosome-like structures with membrane nanotopographies. Single protein tracking shows that, during MAS development, integrins become immobile
and confined within diffusion traps formed by the membrane nanotopographies. Actin filaments also display restricted motion and confinement, indicating strong mechanical connection with
integrins. Using isolated muscle cells, we show that substrate nanotopography, rather than rigidity, drives adhesion maturation by regulating actin protrusion, integrin diffusion and
immobilization. These results thus demonstrate that actin-polymerization-driven membrane protrusions are essential for the formation of strong integrin adhesions sites in the developing
embryo, and highlight the important contribution of geometry to morphogenesis. SIMILAR CONTENT BEING VIEWED BY OTHERS F-ACTIN ARCHITECTURE DETERMINES CONSTRAINTS ON MYOSIN THICK FILAMENT
MOTION Article Open access 16 November 2022 BIOCHEMICAL AND MECHANICAL REGULATION OF ACTIN DYNAMICS Article 02 August 2022 PATTERNED MECHANICAL FEEDBACK ESTABLISHES A GLOBAL MYOSIN GRADIENT
Article Open access 17 November 2022 INTRODUCTION A fundamental question in biology is how to build stable macromolecular structures from highly dynamic proteins. Cell-cell and cell-matrix
adhesions are such structures important for cell migration during tissue morphogenesis, but are also crucial for their mechanical stability1,2. In particular, muscles must be connected by
long-lasting adhesive structures to tendons, tendon cells, or other muscle cells to resist the force of muscle contraction3,4,5. Integrin adhesive structures are composed of a diverse but
evolutionarily conserved set of proteins that link the actin cytoskeleton to the extracellular matrix through transmembrane integrin dimers6,7,8. Actin motion and myosin contractility are
able to generate force on the integrin adhesions through molecular mechanosensors such as talin9 and vinculin10, forming a molecular clutch where actin flow speed and clutch strength are
anti-correlated11,12,13,14. The nanoscale organization and molecular dynamics of integrin adhesions have been well studied in isolated cells, especially on flat non-deformable
substrates15,16,17,18,19. However, cells in vivo reside in a three-dimensional (3D), dynamic, deformable, and confined tissue environment resulting in different molecular organization and
biomechanical control of adhesion formation20,21,22,23. Here we focus on the formation of Drosophila muscle attachment sites (MASs), which are the most prominent integrin adhesive structures
in the developing embryo4. They connect the muscles to specialized epidermal cells, the tendon cells, to anchor their contraction to the apical extracellular matrix. This in vivo model
differs from integrin adhesions formed by cells in culture in fundamental ways. Electron microscopy showed that MASs are not flat structures, but instead are extensively interdigitated24.
This arrangement could increase the strength of the attachment by increasing the surface area of the contact. In addition, membrane molecular organization and nanotopographies can impact the
dynamics and clustering of membrane proteins, including integrins25,26,27,28. Although mechanical force is clearly important for the formation of a fully functional MAS29,30,31; whether
actin polymerization provides that force to an integrin molecular clutch, as is essential in the formation of integrin adhesions on flat surfaces in culture, is not known. Thus, it remains
unclear at the molecular level in a complex 3D tissue environment, how membrane and actin network dynamics regulate the diffusion, immobilization and activation of integrins to trigger the
formation of mechanically stable and mature adhesions. To address these questions, we used an organ culture model of MAS formation compatible with super-resolution microscopy and single
protein tracking. We found that integrins assemble adhesions around Arp2/3-dependent actin protrusions in a structure with nanotopographical patterns where membrane fold into 3D domains
resembling invadosomes. Inhibition of actin protrusions abolished MAS development and disassembled mature MASs. MAS maturation was also accompanied by decreased actin filament movement,
whereas integrin displayed increased molecular confinement and immobilization. We found that these changes in integrin behavior were the result of diffusion traps generated by 3D
nanotopography induced by actin protrusions. Using primary culture of muscle cells, we show that substrate nanotopography, rather than substrate stiffness, promotes the formation of stable
MASs and regulates integrin diffusion and immobilization. These results emphasize the importance of actin-protrusion generated nanotopography in promoting the formation of stable integrin
adhesions in developing tissue and highlight the essential role of geometrical information for morphogenesis. RESULTS SUPER-RESOLUTION MICROSCOPY REVEALS 3D INVADOSOME-LIKE ADHESION
STRUCTURES Examination of the formation of MASs has been hindered by the thickness of the embryo, preventing high spatial and temporal resolution imaging, the morphogenetic movements of the
embryos, preventing long-term tracking of developing MASs, and the vitelline membrane surrounding the embryo, which prevents the straightforward use of inhibitors. Here, we circumvented
these problems by dissecting embryos to produce ex vivo embryo fillets that can be cultured for several hours of MAS development32. We dissected early stage 16 embryos in culture medium, and
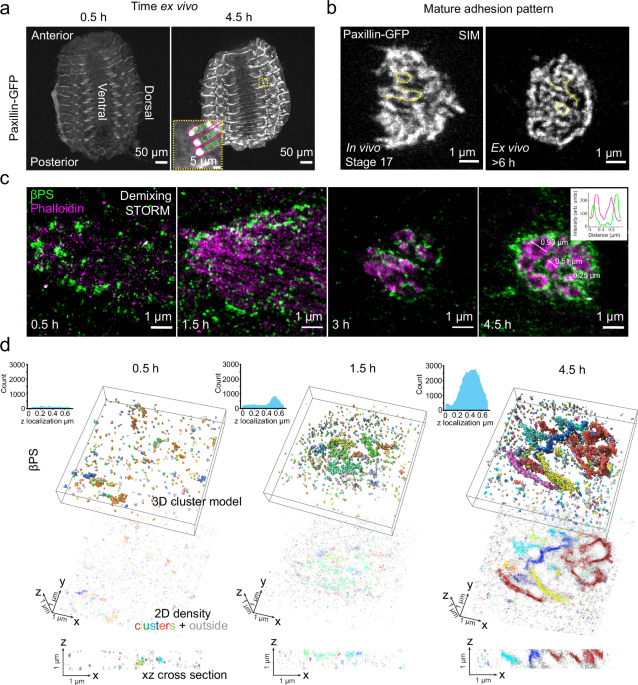
attached the apical surface of the epidermis to the coverslip (Fig. 1a, also See _Methods_ & Supplementary Fig. 1a–d). The main advantage of this is the proximity to the coverslip and
the immobilization of the tissue until the muscles become highly contractile. Importantly, the MASs continue their maturation (Fig. 1a, b and Supplementary Movie 1, 2), the muscles develop
spontaneous contractions normally (Supplementary Movie 3, 4), and embryo fillets show minimal cell death 8 h after dissection (Supplementary Fig. 1i). We focused on the MASs formed by the
lateral-transverse body wall muscles (LT1-3), which are disc-shaped adhesions parallel to the coverslip, and about 5-10 µm away (Fig. 1a and Supplementary Fig. 1b, c). Time-lapse imaging of
developing embryo fillets showed nascent adhesions with isolated dot-like morphology, which then transformed into mature adhesions with enrichment of integrin (βPS-GFP) and paxillin
(paxillin-GFP) (Supplementary Fig. 1e–f and Supplementary Movie 2), within an area of 17.9 ± 0.6 µm2 (mean ± s.e.m., 4.5 h ex vivo) (Supplementary Fig. 1g). The cultured embryo fillets
allowed us to perform quantitative super-resolution microscopy and single protein tracking, and to use pharmacological treatments that target the acto-myosin cytoskeleton and integrins. 2D
and 3D structured illumination microscopy (SIM) imaging of mature adhesion sites in late-stage embryo fillets, and in intact stage 17 embryos, revealed that integrins are patterned into
ridge-like structures, resembling a fingerprint (Fig. 1b, Supplementary Fig. 1h, and Supplementary Fig. 2a, b). Two-color super-resolution imaging with spectral demixing stochastic optical
reconstruction microscopy (STORM) further unveiled the nanoscale organization of MASs and actin. Early MASs exhibit isolated clusters of integrins that, as the MASs mature, become organized
into ridge-shaped structures, with integrin clusters surrounding an actin core (Fig. 1c and Supplementary Fig. 2a, b). These are reminiscent of invadosome-like structures21,33. Other
integrin adhesion proteins, such as talin and vinculin, also present similar patterns in mature MASs (Supplementary Fig.2c, d). With ModLoc 3D STORM super-resolution imaging34 of different
stages of MAS formation, we also observed the transition of integrins from isolated clusters to more elongated structures within the convoluted membrane that extends in the z direction (Fig.
1d, Supplementary Fig. 2e and Supplementary Movie 5). These findings prompted us to examine the role of actin in the formation of this invadosome-like structure, and whether integrin
adhesion proteins display altered mobility and stability within the ridges. ARP2/3-DEPENDENT ACTIN PROTRUSIONS DRIVE ADHESION DEVELOPMENT IN MAS Mechanical forces generated by acto-myosin
contraction and actin flow are essential for integrin adhesion initiation and maturation in cells in culture14,35,36,37. In the MAS, although muscle and non-muscle myosin generate forces
that regulate adhesion reinforcement and protein turn-over29,31, neither is required for the initial development of the adhesion before muscle contraction31,38,39. Consistent with these
findings, when we inhibited Rho-kinase activity with Y-27632 in the embryo fillet to inhibit myosin II contractility, the amount of integrin was reduced but the MASs still matured and formed
fingerprint patterns (Supplementary Fig. 3a, b). This shows that, unlike adhesions in migrating cells in culture, non-muscle myosin contractility is not essential for MAS development and
maintenance but contributes to its force-dependent reinforcement. We then tested whether the force from actin polymerization is important in this system, which has not been addressed with
any genetic or imaging approaches. In cells in culture, the connection of integrins to the retrograde actin flow in cell protrusions, such as the lamellipodium, initiates early
adhesions13,40. This flow is driven by the polymerization of branched actin networks, initiated by the Arp2/3 complex (Arp2/3)41,42. However, mature focal adhesions are connected to stress
fibers composed of unbranched actin filaments, and the elongation of the actin fibers depends on formins instead of Arp2/343. We found that inhibition of Arp2/3 by continuous CK666 treatment
completely abolished MAS maturation (Supplementary Fig. 3c, d), and disassembled mature MASs (Fig. 2a, b and Supplementary Movie. 6). In contrast, treatment with the formin inhibitor SMIFH2
did not affect MAS maturation (Supplementary Fig. 3a, b). Live imaging (every 10 min) throughout MAS formation revealed small actin protrusions in nascent adhesions, which transformed into
more pronounced and stable protrusions in mature adhesions (Fig. 2c–e, Supplementary Fig. 3e, and Supplementary Movie. 7). These actin protrusions may trigger integrin movement and
reorganization observed during MAS formation (Supplementary Fig. 3f and Supplementary Movie 8). Stabilizing actin filaments by Jasplakinolide also prevented adhesion maturation
(Supplementary Fig. 3a, b), supporting the role of dynamic actin protrusions in MAS development. The growth and pattern formation of the MAS thus appears to be driven by dynamic actin
protrusions that recruit integrin around them (Fig. 2f). Using higher temporal resolution imaging (0.1 Hz) at different stages of MAS formation, we also observed highly dynamic actin puncta
and integrin foci in nascent adhesions (1.5 h ex vivo) (Fig. 2g, j) and the formation of more stable structures in mature adhesions (4.5 h ex vivo) (Supplementary Fig. 4a, b and
Supplementary Movie 9). The speed of both actin puncta and integrin foci decreased during MAS development (Supplementary Fig. 4c, d). Actin puncta transitioned from highly dynamic to less
dense but more stable structures, lasting 27.7 ± 0.3 s at 1.5 h to 49.8 ± 1.0 s at 4.5 h (mean ± s.e.m.) (Supplementary Fig. 4e, f). The actin puncta are formed by bursts of actin
polymerization, since their speed and density were reduced by CK666 treatment (Fig. 2g–i and Supplementary Movie 10). Consistent with this, expression of the Arp2/3 complex component
Arp3-GFP, or a component of the WAVE regulatory complex, WAVE-eGFP44,45, in the muscles, showed similar dynamic puncta within developing adhesions (Supplementary Fig. 4g–i). CK666 treatment
also reduced the speed and density of integrin foci (Fig. 2j–l and Supplementary Movie 11). Tracking the movement of actin puncta and integrin foci, to classify moving and stationary
structures (see _Methods_), showed that CK666 treatment similarly increased their stationary fraction compared to moving ones (Supplementary Fig. 4j–l). Altogether, the coordinated assembly
and movement of Arp2/3-dependent actin puncta and integrin foci suggest mechanical coupling between the two, and the reduction of movement speed during adhesion development indicates the
enhancement of their molecular connection13,46. DEVELOPING MASS REGULATE INTEGRIN ADHESION PROTEIN DIFFUSION AND IMMOBILIZATION Membrane nanotopography generated by the actin cytoskeleton
and large membrane glycoproteins can affect membrane protein diffusion and clustering25,27,47. We thus investigated whether the formation of the fingerprint ridges alters integrin diffusion.
We performed high-frequency (50 Hz) single protein tracking coupled with photo-activation localization microscopy (sptPALM), at 1.5 h, 3 h, and 4.5 h (Supplementary Movie 12, see _Methods_
for detail). We used mean square displacement (MSD) analysis to quantify protein diffusive properties including diffusion mode and diffusion coefficient (D)7,17,18 (Fig. 3a–e and
Supplementary Fig. 5a–e). As observed in mature integrin adhesions in cultured cells17,18,48, a significant fraction of integrins (βPS-mEos3.2) remained diffusive inside the adhesion during
its development, and the fraction decreased with time (Fig. 3a, d; 73.7 ± 1.6% at 1.5 h, 49.0 ± 1.3% at 4.5 h, mean ± s.e.m.). Thus, integrin immobilization increased with MAS maturation in
contrast to membrane targeted mEos3.2-CAAX, which continued to diffuse freely inside the MAS with minimal immobilization (Fig. 3b, d, Supplementary Fig. 5i, j, and Supplementary Fig. 6a–d).
Integrin diffusion coefficient decreased during adhesion maturation (0.117 ± 0.003 µm2/s at 1.5 h, 0.084 ± 0.004 µm2/s at 4.5 h, mean ± s.e.m.) (Fig. 3e and Supplementary Fig. 5d, e). The
diffusion coefficient of mEos3.2-CAAX, though higher than that of integrin, also decreased during adhesion development (0.383 ± 0.011µm2/s at 1.5 h, 0.265 ± 0.006 µm2/s at 4.5 h, mean ±
s.e.m.) (Fig. 3e and Supplementary Fig. 6c, d). These results suggest that the increased efficiency of integrin immobilization in MAS is correlated with the formation of the fingerprint
ridges and change in diffusion dynamics. Importantly, the integrin activator kindlin (Fermitin1/Fit1 in _Drosophila_) exhibited different diffusive behavior inside developing MAS (Fig. 3c
and Supplementary Fig. 7a–e). Whereas βPS-mEos3.2 showed increased immobilization, Fit1-mEos3.2 maintained a steady level of immobilization during adhesion development (Fig. 3d).
Furthermore, whereas the two membrane inserted molecules, βPS-mEos3.2 and mEos3.2-CAAX, showed decreased diffusion coefficient and increased confinement, this was not the case for
Fit1-mEos3.2 which showed an increase of diffusion coefficient (Fig. 3e). This suggests the existence of diffusion barriers in the developing MAS restricting the movement of membrane
proteins, while the transient interactions of kindlin with the membrane, via its PH domain binding to PIP218, allows a more dynamic switch between cytosolic and membrane diffusion (Fig. 3f).
DIFFUSION NANO-DOMAINS CONFINE ADHESION PROTEIN MOVEMENT The organization of adhesion into ridge-like structures and restricted diffusion of integrin suggest the formation of diffusion
nano-domains inside MAS (Fig. 3g). Indeed, within mature adhesions, we observed distinct zones with diffusing integrins displaying lower diffusion coefficient and step size, which
corresponded to regions of high paxillin-GFP intensity and increased integrin immobilization (Fig. 3h and Supplementary Fig. 5f). Thus, the MAS is segregated into nano-domains of rapid
versus restricted diffusion. To quantify the confinement of integrin, we calculated the step angle anisotropy49 (see _Methods_) of diffusing βPS-mEos3.2 tracks, which exhibited highly
anisotropic distribution of step angles, compared to mEos3.2-CAAX, indicating a confined behavior (Fig. 3i, j, Supplementary Fig. 5g, h and Supplementary Fig. 6e, f). The step angle
anisotropy of both βPS-mEos3.2 and mEos3.2-CAAX increased during MAS development (Fig. 3j), suggesting increasing geometrical confinement. The anisotropy index increased with larger step
size (>400 nm), comparable to the size of the ridges (Fig. 1c), compared to smaller step size (100 nm) for both βPS-mEos3.2 and mEos3.2-CAAX (Supplementary Fig. 5h and Supplementary Fig.
6f). This suggests that the effect of confinement is due to the geometry of membrane nano-domains within MAS. Different from integrins, the confinement of kindlin does not change during MAS
maturation (Supplementary Fig. 7e), suggesting it can bypass membrane geometrical confinement with cytosolic diffusion (Fig. 3f). In the cell body, where the protrusive structures are not
present, integrins diffuse faster (0.250 ± 0.003 µm2/s, mean ± s.e.m.) with decreased immobilization fraction (27.3 ± 1.6 %, mean ± s.e.m.) and reduced confinement compared to inside MAS
(Supplementary Fig. 8a–j), suggesting that the restricted diffusion dynamics of integrin is specific to the MAS. Altogether, these results indicate that actin protrusions and the formation
of ridge-like structures during MAS maturation could serve as diffusion traps that concentrate membrane proteins and promote their interaction, immobilization, and clustering (Fig.
3f)50,51,52. CONFINED ACTIN MOVEMENT IN MAS FORMS A STRONG CONNECTION WITH INTEGRIN ADHESION The increased immobilization of integrin during adhesion maturation also suggests stronger
engagement with the actin cytoskeleton17,46,53. We thus investigated actin filaments (F-actin) slow movement with low frequency (2 Hz) sptPALM17,54 of actin-mEos3.2 inside MAS (Fig. 4a and
Supplementary Fig. 9a). We first distinguished moving and stationary F-actin and observed an increase of the stationary fraction during MAS development (Fig. 4b). In addition, the moving
fraction displayed isotropic distribution of angles (Fig. 4a and Supplementary Fig. 9b), with mainly highly confined movements, other than directed or Brownian motion (Fig. 4c, d).
Furthermore, the speed of F-actin movement decreased during MAS development (Fig. 4e). CK666 treatment also led to a reduction of F-actin movement (Fig. 4e and Supplementary Fig. 9c, d).
These results are in contrast to the coordinated retrograde F-actin flow observed in the lamellipodium, site of adhesion initiation, and mature integrin adhesions of migrating
cells13,17,40,55,56. To probe the activation status of integrin during MAS development, we performed Mn2+ stimulation to activate integrins17,57,58,59,60 at different stages (Supplementary
Fig. 10a–c) and observed increased immobile fraction in nascent adhesions but not in mature ones (Fig. 4f and Supplementary Fig. 10d), suggesting increased level of integrin activation
during MAS development. As expected for diffusing integrins, only minor effects were seen on the diffusion coefficient and confinement after Mn2+ stimulation (Supplementary Fig. 10e–g). The
increasing fraction of stationary F-actin and decreasing rate of F-actin movement during MAS development, together with increased integrin immobilization and activation, is consistent with
the formation of a stable molecular connection during adhesion maturation (Fig. 4g). DYNAMIC ACTIN PROTRUSIONS AND STABLE ACTIN STRUCTURES REGULATE INTEGRIN DIFFUSION DURING MAS DEVELOPMENT
We then tested whether integrin diffusion and immobilization are controlled by F-actin networks in MASs. Continuous incubation with Latrunculin A or CK666 abolished the maturation of MASs,
decreased the immobilization and confinement of integrin, and increased its diffusion coefficient (Fig. 5a–d, Supplementary Fig. 11a–i and Supplementary Fig. 12a–f). This was specific to the
MAS, unlike the cell body devoid of actin protrusions where integrin diffusion was unaffected by CK666 treatment (Supplementary Fig. 13a–h). Changes in integrin diffusive behavior triggered
by Arp2/3 inhibition were only partially rescued by Mn2+ stimulation (Fig. 5b–d and Supplementary Fig. 12b–f), indicating that direct actin-dependent integrin activation cannot solely
account for the effects on integrin diffusion regulation. Next, we tested whether F-actin networks form a stable structural scaffold in the MAS that could impose geometric constraint on the
membrane and be responsible for the changes in integrin diffusion. We distinguished dynamic from stable actin by performing brief or continuous drug inhibition in nascent and mature MASs
(Fig. 5e, Supplementary Fig. 14a–g and Supplementary Fig. 15a–f). The effect of brief Arp2/3 inhibition on nascent (1.5 h) MASs was quantified 15 min after CK666 treatment (Supplementary
Fig. 14a). This reduced the diffusive fraction of integrins, increased their diffusion coefficient and reduced integrin confinement (Supplementary Fig. 14b–g). However, when mature adhesions
(4.5 h) were briefly treated with CK666, MAS morphology was unaffected (Fig. 5f and Supplementary Fig. 15a), and the immobile fraction was unaltered (Fig. 5h). In contrast to brief CK666
treatment, continuous inhibition of Arp2/3 in mature adhesions led to their disassembly (Fig. 2a, b). Thus, Arp2/3-induced actin polymerization is responsible for a dynamic pool of actin
filament (Fig. 2c-f), both driving membrane protrusions especially in nascent adhesions and the maintenance of a more stable actin structures in mature adhesions. On the other hand, when
mature MAS were treated with Swinholide A, which severs actin filaments, the adhesion structure was immediately perturbed (Fig. 5g, Supplementary Fig. 15b and Movie. 13), resulting in
increased diffusive fraction and diffusion coefficient of integrin (Fig. 5h, i and Supplementary Fig. 15c–e). Together, these results indicate that the coordination of dynamic actin
polymerization and more stable F-actin structures build a 3D scaffold controlling integrin diffusion and immobilization during MAS formation. NANOTOPOGRAPHY PROMOTES ADHESION MATURATION BY
REGULATING ACTIN PROTRUSION AND INTEGRIN DIFFUSION The surface nanotopography of the substrate and its curvature can regulate integrin adhesion and actin dynamics25,61,62. In addition,
substrate stiffness is also an important parameter controlling engagement of the integrin molecular clutch and adhesion formation63,64,65,66. We thus hypothesized that the restriction of
integrin diffusion induced by actin protrusions in the deformable tissue may be a result of its physical properties such as rigidity and topography. In order to test this hypothesis, we
isolated and cultured muscles from mid-stage 16 embryo fillets and placed them on substrates with different biophysical properties. On rigid and flat substrates coated with fibronectin,
muscles only formed nascent dot-like adhesions that did not mature (Fig. 6a). Reducing substrate stiffness did not promote the formation of mature MASs, as isolated muscles only formed
nascent adhesions on polydimethylsiloxane (PDMS) gels with stiffnesses of 3 kPa, 15 kPa and 35 kPa (Supplementary Fig. 16a). The intensity of βPS-GFP even decreased on softer substrates
(Supplementary Fig. 16b), consistent with studies of mechanosensitive integrin adhesions in cultured cells64,65,66. These nascent adhesions were not able to withstand contractile forces, as
the muscles detached during spontaneous muscle contractions (Supplementary Fig. 16c and Supplementary Movie. 14). The reduced adhesion maturation was not due to lower expression of integrin
on soft gels, as we observed that adhesions could still form when muscles contacted each other (Supplementary Fig. 16d). Thus, the rigidity of the muscle-tendon interface is not a
determinant factor of MAS maturation and stabilization. We then plated the muscles on engineered substrates with nanotopographies (600 nm height, 800 nm width) that mimic the geometry
observed in mature MASs. This strongly promoted adhesion maturation and substantially increased the intensity of βPS-GFP, as well as actin, inside adhesions (Fig. 6b, Supplementary Fig. 17a,
b). The adhesions preferentially developed inside the nano-channels with higher intensity on the side walls (Fig. 6b and Supplementary Fig. 17c, d), but also formed clusters on the top and
bottom surfaces (Supplementary Fig. 17e). Using nanotopographies with a fixed depth (600 nm) and varying widths, we observed a size-dependent promotion of adhesion formation on
nano-structured substrates (200 nm, 400 nm, and 800 nm wide) compared to micro-structured (1600 nm wide) and flat substrates (Fig. 6c–e). Thus, the ability to make protrusive structures
confined within nanotopographies is essential for MAS maturation. Since MAS maturation is correlated with the formation of ridge-like structures that could constitute diffusion traps, we
wondered whether nanotopography affects the diffusive behavior of integrin. Tracking of βPS-mEos3.2 showed increased immobilization, decreased diffusion coefficient and increased confinement
on nanopatterns compared to flat substrates (Supplementary Fig. 17e–i). The effects were more pronounced on the bottom surface compared to the top of the ridges (Supplementary Fig. 17g–i),
suggesting that the confinement of integrin diffusion is primarily an effect of the protrusive nano-domains consisted of membrane enveloped adhesion complex and actin cytoskeleton.
mEos3.2-CAAX also displayed a decreased diffusion coefficient and increased confinement on nanopatterns compared to flat substrates. However, in contrast to βPS-mEos3.2, mEos3.2-CAAX showed
similar levels of diffusion coefficient and confinement at the top and bottom surfaces of the nanopattern (Supplementary Fig. 18a–d), suggesting that the actin induced confinement inside the
nano-domains is specific to integrins. CK666 treatment reduced paxillin-GFP intensity on the nanopattern (Supplementary Fig. 19a, b), decreased βPS-mEos3.2 immobile fraction, increased its
diffusion coefficient, and decreased its confinement (Fig. 6f–m and Supplementary Fig. 19c–n), showing that the regulation of integrin diffusion by the nanotopography relies on
Arp2/3-dependent actin polymerization, as in live tissue. Altogether these results suggest that the nanotopography of membrane and the actin cytoskeleton are essential for building
long-lasting integrin adhesions in 3D tissue. DISCUSSION We demonstrate at the molecular level how membrane nanotopography induced by actin-based protrusions can directly influence the
diffusion, immobilization, activation and mechanical connection of adhesion proteins and promote the formation of stable force resisting adhesions in developing tissue (Supplementary Fig.
20a). The nanotopography-induced diffusion trap provides a framework for local regulation of supramolecular organization that promote adhesion proteins aggregation through molecular crowding
and increased probability for protein interactions67. This complements and extends the molecular-clutch-based model of integrin adhesion46,68 in a 3D tissue context where the motion of
adhesion proteins and actin are affected by increased geometrical confinement. We envision the same principle can be applied to diverse 3D cellular and tissue structures in building stable
force-resistant nano-domains that regulate protein diffusion, such as cadherin-mediated cell-cell adhesions and neuromuscular-junctions. Super-resolution microscopy revealed an intricate
fingerprint-like pattern in developing MAS, with submicron sized ridges extending from the muscle surface, which contain an actin core surrounded by integrin clusters. This organization,
with an Arp2/3 actin-rich center surrounded by an integrin adhesion ring, is reminiscent of invadosomes, including podosomes found in various cell types such as monocytes69 and
macrophages70, and invadopodia in cancer cells71. Notably, myoblast fusion also depends on podosome-like structures33. Although MAS adhesions present structurally similar organization of
integrin and actin regulators compared to invadosomes, one major difference is that MASs need to establish stable adhesions to the matrix whereas podosomes and invadosomes usually degrade
the extra cellular matrix (ECM) by secreting metalloproteinases72. On the other hand, MAS adhesions are also structurally different from stable integrin adhesions such as mature focal
adhesions in migrating cells. Mature MASs resist myosin II inhibition but are disassembled by Arp2/3 inhibition. Thus, these findings underline the diversity of integrin adhesions in
different systems and illustrate the importance in studying adhesion structures in tissue environments. Although the importance of substrate and membrane nanotopography in promoting cell
adhesion73, migration74 and differentiation75 has been demonstrated, the molecular and biomechanical mechanisms are not clear. Membrane invaginations recruit actin regulators, including
Arp2/3, BAR domain proteins, and integrins28,62,76, and could mediate adhesion to protein fibers28. Here we provide evidence for the regulation of integrin adhesion by Arp2/3-induced
membrane protrusions. The constraints exerted by membrane curvature26 and membrane proximal actin cytoskeleton77 can limit the diffusion of membrane proteins and promote their clustering27.
The reduction of integrin diffusion and confinement inside diffusion nano-domains, observed in MASs, may help to concentrate adhesion proteins in a dynamic pool where a large portion of
proteins remains diffusive but can be immobilized rapidly17,78. This is consistent with the observation that a small fraction of talin molecules bear force in vivo79, despite the fact that
integrins and associated proteins are very concentrated in the adhesion. The confined movement of actin filaments and restricted diffusion of integrin adhesion proteins potentially create
more possibilities for interaction and may lead to a stronger molecular connection in vivo. Intriguingly, kindlin immobilization and diffusion is apparently not affected by the diffusion
barriers presented in developing adhesions albeit being able to engage in membrane diffusion with its PH domain18, suggesting that kindlin is more dynamic in mature adhesions. This is in
line with the observation that kindlin association with integrin is more transient than talin18. Recent study also shows that the formation of integrin, talin, kindlin ternary complex
increases talin-integrin affinity but reduces kindlin-integrin affinity80. This could potentially explain the paradoxical result of kindlin diffusion and immobilization during MAS
development (Fig. 3d, e) due to its dissociation from the adhesion complex. The ability of kindlin to bypass membrane diffusion barriers via cytosolic diffusion in mature adhesions could
help it rapidly activate integrins elsewhere. In addition to regulating the molecular dynamics of integrins, the organization of the actin cytoskeleton and adhesive structure as well as the
formation of nanotopographic patterns also provide an efficient configuration for force generation and mechanical stability. As the actin networks are sandwiched in between integrin
adhesions, compressive force may help their engagement, thus limiting the optimal size of the nanodomains (Fig. 6c–e) to the submicron regime. In addition, the anchoring of actin protrusion
on both sides, also promotes more efficient protrusions against apposing tissues, which in turn can exert force on the growing adhesions, thus forming a positive feedback loop. This
configuration also results in force generated nearly parallel to the membrane which may help orient actin and adhesion proteins such as talin for mechanotransduction81,82,83. The development
of MAS is also accompanied by secretion and reorganization of the ECM. However, fabricated substrates with controlled ECM coating show the effect of nanotopography in promoting adhesion
formation and actin protrusion does not depend on ECM composition. Nevertheless, in vivo, the organization of ECM could also affect the topography of the adhesion and diffusion of integrin
adhesion proteins, which awaits further study. In-depth analyses of molecular dynamics of integrin-based adhesion were mainly performed using cultured cells adhering to rigid flat
substrates, or on engineered substrates with controlled extensibility and rigidity. Whether insights gained from these model systems could be translated into an understanding of in vivo
processes is still debatable. By dissecting integrin adhesion formation in a physiologically relevant model system that can undergo robust morphogenesis to create force-resistant tissue, our
study help bridge this gap in our current molecular understanding of integrin-mediated adhesion and mechanotransduction. METHODS FLY GENETICS Genomic rescue constructs encoding
Paxillin-GFP84 and Mys-GFP82 were used to mark the muscle attachment sites. These flies were combined with _UAS::Utrophin-GFP_, _UAS::Arp3-GFP_, _UAS::eWAVE-GFP_ and with new constructs made
for this study, as described below: _mys-mEos3.2_, _Fit1-mEos3.2_, _UASp::mEos3.2_, _UASp::mEos3.2-CAAX_, and _UASp::mEos3.2-Actin5C_. The UAS constructs were expressed in the muscle by
crossing to the driver _Mef2::Gal4_. See Supplementary Table 1 in the Supplementary Information for a list of genotypes of fly stocks. TRANSGENIC CONSTRUCTS An insertion of mEos3.2 into the
endogenous locus encoding βPS, _(mys::mEos3.2)_ was generated by homologous recombination, following the same procedure as for _mys-mCherry_85. _Fit1-mEos3.2_ is a genomic rescue construct
of the _Fit1_ gene, tagged with mEos3.2 internally within the F1 loop, with a linker of 4 serine residues at each junction. Thus, setting the A of the initiator ATG as 1, the construct
extends from −1447 to 3994, with mEos3.2 inserted between bases 681 and 682 (amino acids 227 and 228). In a single copy this construct is able to restore viability and fertility to animals
homozygous for a null allele of _Fit1_. The insertion of the fluorescent protein internally within _Fit1_ is essential, as constructs tagged at either the N- or C-terminus of _Fit1_ did not
rescue the null _Fit1_ mutant (unpublished observations). mEos3.2 was amplified from pmEos3.2-C1 plasmid with primers that added NotI and BamHI restriction sites to flank the coding
sequence. The Kozak consensus sequence (CAAA), optimal for translation in _Drosophila_, was included just before the methionine initiation codon of mEos3.2. The CAAX sequence was added into
the C-Terminus of mEos3.2 sequence by PCR amplification using a specific reverse primer that added the CAAX region just before the stop codon amino acid of mEos3.2. The ubiquitous
cytoplasmic actin, encoded by _Actin5C_, was tagged with mEos3.2 at the N-terminus as previously described86. The coding sequence of mEos3.2 was amplifying by PCR and inserted into a UASp
AttP vector, and the amplified Actin5C coding sequence appended to the mEos3.2 sequence with a GSGS linker sequence. See Supplementary Table 2 in the Supplementary Information for a list of
oligonucleotides used in generating transgenic constructs. GENERATION OF TRANSGENIC FLIES _Fit1mEos3.2_, _UAS::mEos3.2_, _UAS::mEos3.2CAAX_ and _UAS::mEos3.2Actin5C_ were inserted into
embryos carrying an AttP docking site at cytological position 51D on the second chromosome, and on the X chromosome the φC31 integrase driven by the vasa promoter to express it within the
germ line. Transformants were selected in the F1 generation by the _w+_ eye color, crosses were performed to remove the integrase and balance the stock. The new UAS lines were validated by
combining with _Mef2::Gal4_ lines to drive expression in muscles and confirming photoconversion of mEos3.2. EMBRYO FIXATION AND IMMUNOFLUORESCENCE MICROSCOPY Embryos were collected on
apple-juice agar plates spotted with fresh yeast. They were aged to stage 15–17 of development and processed for immunofluorescence using standard procedures. βPS-GFP and Vinculin-GFP
embryos and first instar larvae were either imaged live or fixed. The sample were fixed in 4% formaldehyde and stained with phalloidin in PBST (PBS plus 0.5% bovine serum albumin and 0.3%
Triton X-100). βPS-GFP was visualized directly in vivo or in fixed tissue. Comparison with the integrin signal was done using the anti-βPS monoclonal antibody (CF.6G11, Developmental studies
hybridoma bank). Actin visualization was performed using Rhodamine phalloidin and Alexa Fluor 647 phalloidin (Life Technologies) used at 1:500 and 1:50, respectively. Samples were scanned
with a Leica SP8 confocal microscope using a 20X/0.75 NA objective for whole-embryo imaging or a 60X/1.40NA Oil objective for muscle attachments. SIM AND LIGHTNING MICROSCOPY IN WHOLE
EMBRYOS 3D-SIM super-resolution images were performed using a Zeiss Elyra 7 Lattice SIM, the system equipped with two sCMOS cameras allows the 2-color acquisition. The images were capture
with 63X/1.4NA oil immersion objective. Raw data was reconstructed using ZEN software. Lightning microscopy used to obtain Vinculin-GFP high-resolution images were perform with Leica
Stellaris 5 confocal microscope using 63X/1.4NA oil immersion objective. Images were acquired with the optimal Z-step size covering several LT1-3 adhesion sites from the same embryo of a
total of 22 wild type embryos. 3D visualization was done using Fiji software from a region of interest of 8-10 µm2 to cover a whole adhesion site. EMBRYO FILLET DISSECTION Flies were allowed
to lay eggs on a grape juice agar plate at 25 °C for 2 h and then aged at 18–22 °C. Early stage 16 embryos were selected based on gut morphology (to synchronize the timing of adhesion
formation we started dissection at the point when the constrictions of the midgut segments begin to lose left-right symmetry). Embryos were briefly washed with 70% Ethanol and MQ water
before dissection. The chorion was removed by rolling the embryo on a double-sided tape. During the dissection, a series of cuts were made with a tungsten needle along the mid-dorsal line
from the posterior to the anterior that penetrate the vitelline membrane and epithelium while avoiding damaging the musculature. Embryos were then transferred and adhered to a 0.1%
poly-L-lysine (Sigma) coated coverslip and flattened to form a fillet. The gut was removed to reduce autofluorescence and to prevent contractions that could interfere with imaging. Embryo
fillets were washed 3 times with PBS and allowed to develop in Shields and Sang M3 insect medium (Sigma) supplemented with 2% FBS, 0.01 mg/ml Insulin (Sigma) and Pen/Strep. EMBRYO FILLET
FIXATION AND IMMUNOSTAINING Embryo fillets were fixed in PEM buffer (80 mM PIPES, 5 mM EGTA, 2 mM MgCl2, pH 6.8) with 4% paraformaldehyde and 0.2% glutaraldehyde for 10 min at 37 °C.
Subsequently, the embryo fillets were washed 3 times with PBS and permeabilized in PBST (0.2% Triton X-100) for 5–7 days at 4 °C, before blocking in PBST with 3% BSA overnight at 4 °C. The
following primary and secondary antibodies was used: anti-βPS monoclonal antibody (CG6G11, Developmental studies hybridoma bank) (1:15), CF680 goat anti-mouse IgG (Biotium, Cat: 20817-500
µl, Lot: 20C0827) (1:200). F-actin was visualized with phalloidin-AF647 (Life Technologies) (1:100). CONFOCAL AND SIM MICROSCOPY OF LIVE EMBRYO FILLET Confocal and SIM acquisitions were
performed on a spinning disk confocal microscope (Leica DMI8, 100 × 1.49 N.A. oil immersion objective, sCMOS Camera Photometrics Prime 95B). For MAS development over several hours, one
z-stack of 10 μm with 0.2 μm step size was acquired at 10 min per frame for 6–8 h. For fast dynamics of integrin clusters and actin puncta, one z-stack of 5 μm with 0.2 μm step size was
acquired at 10 s per frame for 5–10 min. Super-resolution SIM images were acquired with the Live-SR module. SPECTRAL DEMIXING STORM Spectral demixing STORM87 acquisition was performed on an
Abbelight SAFe 360 module installed on and inverted microscope (Nikon Ti) with a CFI Apo TIRF 100X oil 1.49 NA objective using HiLo illumination. Samples stained with AF647 and CF680 was
imaged with a 640 nm laser. Fluorescence was collected with a dichroic (Dio02-R561-25 × 36, Semrock) and an emission filter (FF01-432/515/595/730-25, Semrock). The fluorescence was
subsequently separated by a dichroic (FF699-Fdi01-t3-25 × 36, Semrock) and collected by two cameras (Hamamatsu ORCA-Flash 4.0). The images were analyzed with the Abbelight Neo software as
per the manufacturer’s instructions. Demixing of the localizations based on the ratio (0–1) of photons from the two cameras were calculated and the ratios 0–0.4 were assigned to the AF647
channel while the ratios 0.6–1 were assigned to the CF680 channel. Ratios between 0.4 and 0.6 were rejected. 3D MODLOC STORM For 3D ModLoc STORM34, the sample was excited by a 639 nm laser
(Genesis MX 639, 1W, Coherent) and imaged through a Nikon APO TIRF 60 × 1.49 NA oil immersion objective lens mounted on an inverted microscope (Eclipse Ti, Nikon). The fluorescence was
collected with a dichroic (Di03-R635-t1-25 × 36, Semrock) and an emission filter (BLP01-635R-25, Semrock) on four quadrants of a 512 × 512 pixel EMCCD camera (iXon3, Andor). An EOM
(EO-PM-NR-C1, Thorlabs) was used for the phase modulation at the excitation whereas a Pockels cell (CF1043-20SG-500/700, Fast pulse) performed the demodulation at the detection. 60 cycles of
modulation/demodulation were performed during the acquisition time of one image (50 ms). Typically, between 20,000 and 30,000 images were acquired to reconstruct the 3D super-resolved
images. Drosophila embryo fillet samples were imaged 5–10 μm in depth with a z range of 1 μm. SPTPALM Embryo fillets or isolated muscle cells were imaged at 26 °C in a Ludin chamber (Life
Imaging Services) on and inverted microscope (Nikon Ti) with a CFI Apo TIRF 100X oil 1.49 NA objective with HiLo illumination. A 561 nm laser (Cobolt, 20 mW out of objective) was used to
excite photo-converted mEos3.2 while simultaneously photo-converting new molecules with a 405 nm laser (Omicron, 5 mW out of objective). The laser power of the 405 nm laser was adjusted for
each sample to control the density of converted mEos3.2 and to ensure a spatial temporal separation of single molecule signals enabling localization and tracking. Fluorescence of mEos3.2 was
collected with a Di01-R561 dichroic and FF01-617/73 emission filter (Semrock) and an EMCCD camera (Evolve, Photometrics) Image streams were acquired with Metamorph software (Molecular
Devices) at 50 Hz (20 ms per frame) and ~10,000 frames were acquired for each set of experiment. A reference image of Pax-GFP is acquired before and after the stream with a GFP filter cube
(excitation filter: FF01-472/30, dichroic: FF-495Di02, emission filter: FF02-520/28; Semrock). SINGLE MOLECULE TRACKING ANALYSIS Single molecule fluorescence from sptPALM images were
localized with the PALMtracer software88 where a wavelet-based method is used to identify the spots followed by gaussian fitting of the point spread function to localize the 2D position of
the fluorophore. Single molecule localization from consecutive frames are then linked with a maximum distance of 3 pixels per frame to track diffusing molecules. Trajectories lasting at
least 10 frames were processed with a custom written MATLAB code for mean squared displacement (MSD) analysis. The MSD is calculated as: $${MSD}(t=n\cdot \Delta t)=\frac{
{\sum}_{i-1}^{N-n}{({x}_{i+n}-{x}_{i})}^{2}+{({y}_{i+n}-{y}_{i})}^{2}}{N-n}$$ (1) Where xi and yi are the 2D coordinates of the molecule at time \({{\boldsymbol{i}}} \cdot \Delta
{{\boldsymbol{t}}}\). The proteins in the cell can exhibit anomalous diffusion at longer time scale but at shorter time scale will appear diffusive with a close to linear MSD curve. We fit
the first 4 points of the MSD and take the slope as the apparent diffusion coefficient D. In order to differentiate the diffusive from immobile trajectories, we use two parameters Dthresh
and Rconf. Dthresh = (2.355 σ) 2/(4 × 4 × 0.02 s) is the minimum D observable limited by the localization uncertainty σ of the imaging system. Rconf is the confinement radius by fitting the
first 80% of the MSD curve with the following: $${MSD}\left(t\right)=\,\frac{4{r}_{{conf}}^{2}}{3}(1-{e}^{-\frac{t}{\tau }})$$ (2) where τ =
\({{{\boldsymbol{r}}}}_{{{\boldsymbol{conf}}}}^{{{\boldsymbol{2}}}}\)/3\({{{\boldsymbol{D}}}}_{{{\boldsymbol{conf}}}}\). We characterize a trajectory as immobile if its D < Dthresh and
Rconf < 3σ. For each set of experimental conditions, the localization uncertainty σ is characterized with the ThunderSTORM plugin in Fiji, and the resolution is determined as 2.355 σ
(88.3 nm for embryo fillet and 93.8 nm for Curi Bio nano-pattern). The respective Dthresh is 0.0243 µm2/s for embryo fillet and 0.0275 µm2/s for Curi Bio nano-pattern, and is represented by
the gray area in the diffusion coefficient distribution graphs. In order to quantify the confinement of the trajectories, we measure the step angle between consecutive steps of only
diffusive trajectories and only for steps larger than the resolution limit. From the distribution of the angles we then calculate the step angle anisotropy = f150–210/60 × 360, where
f150–210 is the fraction of angles between 150 and 210 degrees. DRUG TREATMENT AND MN2+ STIMULATION Chemical inhibitors were administered at the following concentration: Latrunculin A (Enzo
Life Sciences), 10 mM; CK666 (Tocris), 250 μM; Swinholide A (Abcam), 100 nM; Y-27632 (Sigma), 100 μM; SMIFH2 (Tocris), 10 μM; Jasplakinolide (Tocris), 3 μM. Due to the photo-sensitivity of
Y-27632, the treatment was performed in a dark environment prior to imaging. A brief stimulation with 5 mM Mn2+ was performed in Ringer’s solution (150 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM
MgCl2, 10 mM Hepes and 2 g/L Glucose, pH 7.4) at different developmental stages for 15 min before sptPALM imaging. This was followed by exchanging to culture medium after imaging to allow
embryo fillet development. F-ACTIN MOVEMENT MEASUREMENT Female flies carrying _UAS::Actin-mEos3.2_ and _Paxillin-GFP_ were crossed with males carrying _Mef2::GAL4_. Embryos were collected
and dissected for fillets. Expression of Actin-mEos3.2 was confirmed with photo-activation of 405 laser before imaging. Single molecule dynamics of Actin-mEos3.2 was imaged with 2 Hz frame
rate and 500 ms exposure time with a 561 laser and HiLo illumination. This imaging frequency excludes the fast diffusing actin monomers and tracks only actin within slow moving filaments.
Localization and trajectories linking were performed with PALMtracer. Trajectories were classified into mobile and stationary with MSD analysis using the following parameters: Dthresh =
0.000975 µm2/s, _σ_ = 0.0375 µm. A custom written MATLAB routine was used to calculate the actin movement speed and distribution of angles. The movement speed was defined as the distance
traveled from the beginning to the end of the track divided by duration of track. Movement angle was based on the vector from the beginning of the track to the end of the track. To
characterize the type of movement of actin trajectories, the mobile trajectories were fitted with the power law 4Dtα. Tracks with _α_ < 0.8 were classified as sub diffusive, 0.8 < _α_
< 1.2 were classified as Brownian diffusion and _α_ > 1.2 were classified as directed motion89. ACTIN PUNCTA AND INTEGRIN FOCI TRACKING Adhesions in embryo fillets expressing
_Utrophin-GFP_ or _βPS-GFP_ were imaged at different developmental stages at 10 sec/frame with a spinning disk confocal microscope (Leica DMI8, 100 × 1.49 N.A. oil immersion objective, sCMOS
Camera Photometrics Prime 95B). Actin and integrin images were first background subtracted with a 3-pixel radius rolling ball to reveal dynamic puncta from stable structures. Puncta were
detected and tracked with Fiji TrackMate plugin. Speed of the puncta was calculated as instantaneous speed from consecutive frames. SOFT PDMS GEL FABRICATION AND FUNCTIONALIZATION Soft PDMS
gel base (Dowsil CY-52-276-A) and curing agent (Dowsil CY-52-276-B) were mixed at ratios 1.2:1, 1:1, and 1:1.2 to produce gels with different rigidities of ~3 kPa, 15 kPa and 35 kPa
respectively90. One drop of mixed gel was deposited on an 18 mm coverslip and spin coated at 2000 rpm for 30 s to produce a thin layer of ~30–50 µm. The gels were cured at 80 °C for 2 h. To
functionalize the surface of the gel, 5% (3-aminopropyl) triethoxysilane (APTES) (Sigma) was deposited on the gel in ethanol for 5 min, washed 1X with ethanol and subsequently cured at 70 °C
for 30 min. Then 10 μg/ml Human Fibronectin (Sigma) was coated for 1 h at 37 °C before washing and seeding of cells. FABRICATION OF QUARTZ NANOPATTERN Nanopatterned gratings were fabricated
on cleared fused quartz coverslips measuring 18 mm in diameter and 0.2 ± 0.05 mm in thickness, obtained from Technical Glass Product Inc. Prior to usage, the coverslips underwent a thorough
cleaning process involving acetone and isopropanol with sonication to remove any particles. Subsequently, the cleaned coverslips were blow-dried and heated at 180 degrees on a hotplate to
ensure complete dryness. We then spin-coated a 275 nm layer of 9% CSAR 62 Ebeam resist onto the substrate at 2000 rpm and baked it at 180 degrees for 3 min. On top of this resist layer, we
spin-coated a 100 nm conductive layer of Electra 92 at 1000 rpm and baked it for an additional 3 min. Using the Raith Voyager lithography system, we patterned the gratings with various
dimensions (200 nm, 400 nm, 800 nm, 1600 nm) and pitches, incorporating multiple biases to compensate for proximity effects. Following lithography exposure, the patterned substrate was
immersed in DI water to remove the Electra 92 conductive layer and then developed in xylene for 45 s to reveal the nanopatterns. We evaporated a 50 nm Cr layer onto the substrate as the
masked and performed plasma etching using the PlasmaTherm Versaline LL ICP Dielectric Etcher for 600 nm. The Cr layer was removed with a Cr etchant for 30 min, leaving us with the desired
nanopatterned gratings. ISOLATION OF EMBRYONIC MUSCLE CELLS AND IN VITRO CULTURE Mid stage 16 Drosophila embryos (after symmetry breaking of the gut, roughly corresponding to 3 h ex vivo)
were harvested and dissected as described above to produce a flat embryo fillet attached to a poly-L-lysine (Sigma) coated coverslip. Excess tissue including the gut, CNS and trachea were
removed with a dissection needle. Other isolated cells were removed by gentle pipetting. The remaining musculature and epithelium attached to the cuticle were then washed 3X with PBS. A
short treatment of 37 °C pre-warmed trypsin (without EGTA, Invitrogen) was administered for 3 min at room temperature to loosen the muscle tissue before exchanging to insect medium with 10%
FBS. The muscle cells were then carefully teased out with a dissection needle and transferred with a 20 μl pipette tip to petri dishes with different substrates. The muscle cells were
allowed to attach and develop adhesions for 1.5 h before imaging. For experiments in Fig. 6c–e, a quartz coverslip with fabricated nanopatterns of different sizes were used as substrate. The
surface was treated with oxygen plasma for 5 min before coating with 10 μg/ml Human Fibronectin (Sigma) for 1 h at 37 °C, and overnight at 4 °C. After cell seeding and imaging, the
coverslip was washed with 10% SDS and subsequently immersed in concentrated sulfuric acid for 3 days to remove organic matter before washing with MQ water and reuse. For experiments in Fig.
6a, b, f–m and Supplementary Figs. 17–19, a commercial polymer nano-pattern or flat surface (Curi Bio) was used. IMAGE PROCESSING Fluorescence images displayed in the figures were background
subtracted and adjusted for better contrast. Fluorescence intensity measurements were done using raw images, where camera offset measured from closed shutter images were first subtracted.
ModLoc super-resolution localization data was first processed with a custom Python code to perform DBSCAN (scikit-learn) clustering analysis. Then, localizations outside clusters
(represented by gray dots in Fig. 1d 2D density) were filtered out and 3D surface models were generated with ChimeraX software, a random color is assigned to each cluster. 3D fluorescence
images were rendered with Napari, and movies were generated with the animation plugin with key frames. STATISTICAL ANALYSIS Data presented in bar and line graphs represent mean ± s.e.m.
unless otherwise specified. Data presented in boxplots represent median ± 25/75 percentiles with range and statistical outliers marked. Data exclusion is based on sample size (minimum number
of trajectories >100 per region of interest and length of individual trajectories ≥10) while statistical outliers were not excluded from analysis. Statistical tests were conducted with
MATLAB. Two-tailed student’s _t_-tests were performed to compare data sets with n.s. representing _p_ ≥ 0.05, * representing _p_ < 0.05, ** representing _p_ < 0.01 and *** representing
_p_ < 0.001. Normality of the data was tested by Anderson-Darling test with a confidence interval of 0.05. REPORTING SUMMARY Further information on research design is available in the
Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY The data used this study are provided in the Supplementary Information/Source Data file. Source data are provided
with this paper. CODE AVAILABILITY Custom computer codes used in this study are accessible at Zenodo [https://doi.org/10.5281/zenodo.13773835]. REFERENCES * Lecuit, T., Lenne, P.-F. F.
& Munro, E. Force generation, transmission, and integration during cell and tissue morphogenesis. _Annu. Rev. Cell Dev. Biol._ 27, 157–184 (2011). Article PubMed CAS Google Scholar *
Goodwin, K. et al. Basal cell-extracellular matrix adhesion regulates force transmission during tissue morphogenesis. _Dev. Cell_ 39, 611–625 (2016). Article PubMed CAS Google Scholar *
Subramanian, A. & Schilling, T. F. Tendon development and musculoskeletal assembly: emerging roles for the extracellular matrix. _Development_ 142, 4191–4204 (2015). Article PubMed
CAS PubMed Central Google Scholar * Maartens, A. P. & Brown, N. H. The many faces of cell adhesion during Drosophila muscle development. _Dev. Biol._ 401, 62–74 (2015). Article
PubMed CAS Google Scholar * Valdivia, M., Vega-Macaya, F. & Olguín, P. Mechanical control of myotendinous junction formation and tendon differentiation during development. _Front.
Cell Dev. Biol._ 5, 26 (2017). Article PubMed PubMed Central Google Scholar * Kechagia, J. Z., Ivaska, J. & Roca-Cusachs, P. Integrins as biomechanical sensors of the
microenvironment. _Nat. Rev. Mol. Cell Biol._ 20, 457–473 (2019). Article PubMed CAS Google Scholar * Vicente, F. N., Chen, T., Rossier, O. & Giannone, G. Novel imaging methods and
force probes for molecular mechanobiology of cytoskeleton and adhesion. _Trends Cell Biol._ 33, 204–220 (2023). Article Google Scholar * Kanchanawong, P. & Calderwood, D. A.
Organization, dynamics and mechanoregulation of integrin-mediated cell–ECM adhesions. _Nat. Rev. Mol. Cell Biol._ 24, 142–161 (2023). Article PubMed CAS Google Scholar * Jiang, G.,
Giannone, G., Critchley, D. R., Fukumoto, E. & Sheetz, M. P. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. _Nature_ 424, 334–337 (2003). Article
ADS PubMed CAS Google Scholar * Thievessen, I. et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. _J. Cell
Biol._ 202, 163–177 (2013). * Chan, C. E. & Odde, D. J. Traction dynamics of filopodia on compliant substrates. _Science_ 322, 1687–1691 (2008). Article ADS PubMed CAS Google Scholar
* Jaumouillé, V., Cartagena-Rivera, A. X. & Waterman, C. M. Coupling of β2 integrins to actin by a mechanosensitive molecular clutch drives complement receptor-mediated phagocytosis.
_Nat. Cell Biol._ 21, 1357–1369 (2019). * Gardel, M. L. et al. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. _J. Cell Biol._ 183, 999–1005
(2008). Article PubMed CAS PubMed Central Google Scholar * Hu, K., Ji, L., Applegate, K. T., Danuser, G. & Waterman-Storer, C. M. Differential transmission of actin motion within
focal adhesions. _Science_ 315, 111–115 (2007). Article ADS PubMed CAS Google Scholar * Kanchanawong, P. et al. Nanoscale architecture of integrin-based cell adhesions. _Nature_ 468,
580 (2010). Article ADS PubMed CAS PubMed Central Google Scholar * Stubb, A. et al. Superresolution architecture of cornerstone focal adhesions in human pluripotent stem cells. _Nat.
Commun._ 10, 4756 (2019). Article ADS PubMed PubMed Central Google Scholar * Rossier, O. et al. Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal
adhesions. _Nat. Cell Biol._ 14, 1057–1067 (2012). Article PubMed CAS Google Scholar * Orré, T. et al. Molecular motion and tridimensional nanoscale localization of kindlin control
integrin activation in focal adhesions. _Nat. Commun._ 12, 3104 (2021). Article ADS PubMed PubMed Central Google Scholar * Tsunoyama, T. A. et al. Super-long single-molecule tracking
reveals dynamic-anchorage-induced integrin function. _Nat. Chem. Biol._ 14, 497–506 (2018). Article PubMed CAS Google Scholar * Labernadie, A. et al. Protrusion force microscopy reveals
oscillatory force generation and mechanosensing activity of human macrophage podosomes. _Nat. Commun._ 5, 5343 (2014). Article ADS PubMed CAS Google Scholar * Dries, Kvanden et al.
Modular actin nano-architecture enables podosome protrusion and mechanosensing. _Nat. Commun._ 10, 5171 (2019). Article ADS PubMed PubMed Central Google Scholar * Kuhn, T. et al.
Single-molecule tracking of Nodal and Lefty in live zebrafish embryos supports hindered diffusion model. _Nat. Commun._ 13, 6101 (2022). Article ADS PubMed CAS PubMed Central Google
Scholar * Massou, S. et al. Cell stretching is amplified by active actin remodelling to deform and recruit proteins in mechanosensitive structures. _Nat. Cell Biol._ 22, 1011–1023 (2020).
Article PubMed CAS Google Scholar * Prokop, A., Martı́n-Bermudo, M. D., Bate, M. & Brown, N. H. Absence of PS Integrins or Laminin A Affects Extracellular Adhesion, but Not
Intracellular Assembly, of Hemiadherens and Neuromuscular Junctions in Drosophila Embryos. _Dev. Biol._ 196, 58–76 (1998). Article PubMed CAS Google Scholar * Paszek, M. J. et al. The
cancer glycocalyx mechanically primes integrin-mediated growth and survival. _Nature_ 511, 319–325 (2014). Article ADS PubMed CAS PubMed Central Google Scholar * Reynwar, B. J. et al.
Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. _Nature_ 447, 461–464 (2007). Article ADS PubMed CAS Google Scholar * Saltukoglu, D. et al. Plasma
membrane topography governs the 3D dynamic localization of IgM B cell antigen receptor clusters. _EMBO J._ 42, e112030 (2023). Article PubMed CAS PubMed Central Google Scholar * Zhang,
W. et al. Curved adhesions mediate cell attachment to soft matrix fibres in three dimensions. _Nat. Cell Biol._ 25, 1453–1464 (2023). Article PubMed CAS PubMed Central Google Scholar *
Yuan, L., Fairchild, M. J., Perkins, A. D. & Tanentzapf, G. Analysis of integrin turnover in fly myotendinous junctions. _J. Cell Sci._ 123, 939–946 (2010). Article PubMed CAS Google
Scholar * Pines, M. et al. Mechanical force regulates integrin turnover in Drosophila in vivo. _Nat. Cell Biol._ 14, 935–943 (2012). Article ADS PubMed CAS Google Scholar * Bulgakova,
N. A., Wellmann, J. & Brown, N. H. Diverse integrin adhesion stoichiometries caused by varied actomyosin activity. _R. Soc. Open Biol._ 7, 160250 (2017). Article Google Scholar *
Broadie, K., Skaer, H. & Bate, M. Whole-embryo culture of Drosophila: development of embryonic tissues in vitro. _Rouxs Arch. Dev. Biol._ 201, 364–375 (1992). Article PubMed Google
Scholar * Sens, K. L. et al. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. _J. Cell Biol._ 191, 1013–1027 (2010). Article PubMed CAS PubMed
Central Google Scholar * Jouchet, P. et al. Nanometric axial localization of single fluorescent molecules with modulated excitation. _Nat. Photonics_ 15, 297–304 (2021). Article ADS CAS
Google Scholar * Choi, C. K. et al. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. _Nat. Cell Biol._ 10,
1039–1050 (2008). Article PubMed CAS PubMed Central Google Scholar * Giannone, G. et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. _Cell_ 128,
561–575 (2007). Article PubMed CAS PubMed Central Google Scholar * Plotnikov, S. V., Pasapera, A. M., Sabass, B. & Waterman, C. M. Force fluctuations within focal adhesions mediate
ECM-rigidity sensing to guide directed cell migration. _Cell_ 151, 1513–1527 (2012). Article PubMed CAS Google Scholar * Rui, Y., Bai, J. & Perrimon, N. Sarcomere formation occurs
by the assembly of multiple latent protein complexes. _PLoS Genet._ 6, e1001208 (2010). Article PubMed PubMed Central Google Scholar * Bloor, J. W. & Kiehart, D. P. zipper Nonmuscle
myosin-II functions downstream of PS2 integrin in Drosophila myogenesis and is necessary for myofibril formation. _Dev. Biol._ 239, 215–228 (2001). Article PubMed CAS Google Scholar *
Giannone, G. et al. Periodic lamellipodial contractions correlate with rearward actin waves. _Cell_ 116, 431–443 (2004). Article PubMed CAS Google Scholar * Mullins, R. D., Heuser, J. A.
& Pollard, T. D. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. _Proc. Natl Acad. Sci.
USA_ 95, 6181–6186 (1998). Article ADS PubMed CAS PubMed Central Google Scholar * Svitkina, T. & Borisy, G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic
organization and treadmilling of actin filament array in lamellipodia. _J. Cell Biol._ 145, 1009–1026 (1999). Article PubMed CAS PubMed Central Google Scholar * Valencia, F. R. et al.
Force-dependent activation of actin elongation factor mDia1 protects the cytoskeleton from mechanical damage and promotes stress fiber repair. _Dev. Cell_ 56, 3288–3302.e5 (2021). Article
PubMed CAS Google Scholar * Machesky, L. M. et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. _Proc. Natl Acad. Sci. USA_ 96, 3739–3744
(1999). Article ADS PubMed CAS PubMed Central Google Scholar * Fricke, R. et al. Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE.
_Curr. Biol._ 19, 1429–1437 (2009). Article PubMed CAS Google Scholar * Case, L. B. et al. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal
adhesions. _Nat. Cell Biol._ 17, 880–892 (2015). Article PubMed CAS PubMed Central Google Scholar * Ketchum, C. M. et al. Subcellular topography modulates actin dynamics and signaling
in B-cells. _Mol. Biol. Cell_ 29, 1732–1742 (2018). Article PubMed CAS PubMed Central Google Scholar * Shibata, A. C. et al. Archipelago architecture of the focal adhesion: Membrane
molecules freely enter and exit from the focal adhesion zone. _Cytoskeleton_ 69, 380–392 (2012). Article PubMed CAS Google Scholar * Hansen, A. S., Amitai, A., Cattoglio, C., Tjian, R.
& Darzacq, X. Guided nuclear exploration increases CTCF target search efficiency. _Nat. Chem. Biol._ 16, 257–266 (2020). Article PubMed CAS Google Scholar * Lee, Y. et al.
High-throughput, single-particle tracking reveals nested membrane domains that dictate KRasG12D diffusion and trafficking. _eLife_ 8, e46393 (2019). Article PubMed PubMed Central Google
Scholar * Huang, W. Y. et al. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. _Science_ 363, 1098–1103 (2019). Article ADS PubMed CAS
PubMed Central Google Scholar * Heck, J. et al. Transient confinement of CaV2.1 Ca2+-channel splice variants shapes synaptic short-term plasticity. _Neuron_ 103, 66–79.e12 (2019). Article
PubMed CAS Google Scholar * Nordenfelt, P., Elliott, H. L. & Springer, T. A. Coordinated integrin activation by actin-dependent force during T-cell migration. _Nat. Commun._ 7,
13119 (2016). Article ADS PubMed CAS PubMed Central Google Scholar * Mehidi, A. et al. Forces generated by lamellipodial actin filament elongation regulate the WAVE complex during cell
migration. _Nat. Cell Biol._ 23, 1148–1162 (2021). Article PubMed CAS Google Scholar * Ponti, A., Machacek, M., Gupton, S., Waterman-Storer, C. & Danuser, G. Two distinct actin
networks drive the protrusion of migrating cells. _Science_ 305, 1782–1786 (2004). Article ADS PubMed CAS Google Scholar * Hotulainen, P. & Lappalainen, P. Stress fibers are
generated by two distinct actin assembly mechanisms in motile cells. _J. Cell Biol._ 173, 383–394 (2006). Article PubMed CAS PubMed Central Google Scholar * Luo, B.-H., Springer, T. A.
& Takagi, J. Stabilizing the open conformation of the integrin headpiece with a glycan wedge increases affinity for ligand. _Proc. Natl Acad. Sci. USA_ 100, 2403–2408 (2003). Article
ADS PubMed CAS PubMed Central Google Scholar * Cluzel, C. et al. The mechanisms and dynamics of αvβ3 integrin clustering in living cells. _J. Cell Biol._ 171, 383–392 (2005). Article
PubMed CAS PubMed Central Google Scholar * Bunch, T. A. et al. Amino acid changes in Drosophila αPS2β PS integrins that affect ligand affinity*. _J. Biol. Chem._ 281, 5050–5057 (2006).
Article PubMed CAS Google Scholar * López-Ceballos, P., Herrera-Reyes, A. D., Coombs, D. & Tanentzapf, G. In vivo regulation of integrin turnover by outside-in activation. _J. Cell
Sci._ 129, 2912–2924 (2016). Article PubMed Google Scholar * Li, X. et al. Nanoscale surface topography reduces focal adhesions and cell stiffness by enhancing integrin endocytosis. _Nano
Lett._ 21, 8518–8526 (2021). Article ADS PubMed CAS PubMed Central Google Scholar * Gaertner, F. et al. WASp triggers mechanosensitive actin patches to facilitate immune cell
migration in dense tissues. _Dev. Cell_ 57, 47–62.e9 (2022). Article PubMed CAS PubMed Central Google Scholar * Elosegui-Artola, A. et al. Mechanical regulation of a molecular clutch
defines force transmission and transduction in response to matrix rigidity. _Nat. Cell Biol._ 18, 540–548 (2016). Article PubMed CAS Google Scholar * Pelham, R. J. & Wang, Y. Cell
locomotion and focal adhesions are regulated by substrate flexibility. _Proc. Natl Acad. Sci. USA_ 94, 13661–13665 (1997). Article ADS PubMed CAS PubMed Central Google Scholar *
Ghibaudo, M. et al. Traction forces and rigidity sensing regulate cell functions. _Soft Matter_ 4, 1836–1843 (2008). Article ADS CAS Google Scholar * Ghassemi, S. et al. Cells test
substrate rigidity by local contractions on submicrometer pillars. _Proc. Natl Acad. Sci. USA_ 109, 5328–5333 (2012). Article ADS PubMed CAS PubMed Central Google Scholar * Nawrocki,
G., Wang, P., Yu, I., Sugita, Y. & Feig, M. Slow-down in diffusion in crowded protein solutions correlates with transient cluster formation. _J. Phys. Chem. B_ 121, 11072–11084 (2017).
Article PubMed CAS PubMed Central Google Scholar * Oria, R. et al. Force loading explains spatial sensing of ligands by cells. _Nature_ 552, 219–224 (2017). Article ADS PubMed CAS
Google Scholar * Linder, S. et al. The polarization defect of Wiskott-Aldrich syndrome macrophages is linked to dislocalization of the Arp2/3 complex. _J. Immunol._ 165, 221–225 (2000).
Article PubMed CAS Google Scholar * Linder, S., Nelson, D., Weiss, M. & Aepfelbacher, M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. _Proc.
Natl Acad. Sci. USA_ 96, 9648–9653 (1999). Article ADS PubMed CAS PubMed Central Google Scholar * Gligorijevic, B. et al. N-WASP-mediated invadopodium formation is involved in
intravasation and lung metastasis of mammary tumors. _J. Cell Sci._ 125, 724–734 (2012). Article PubMed CAS PubMed Central Google Scholar * Linder, S. The matrix corroded: podosomes and
invadopodia in extracellular matrix degradation. _Trends Cell Biol._ 17, 107–117 (2007). Article PubMed CAS Google Scholar * Yim, E. K. F., Darling, E. M., Kulangara, K., Guilak, F.
& Leong, K. W. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. _Biomaterials_ 31, 1299–1306
(2010). Article PubMed CAS Google Scholar * Reversat, A. et al. Cellular locomotion using environmental topography. _Nature_ 582, 582–585 (2020). Article ADS PubMed CAS Google
Scholar * Yim, E. K. F., Pang, S. W. & Leong, K. W. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. _Exp. Cell Res._ 313,
1820–1829 (2007). Article PubMed CAS PubMed Central Google Scholar * Lou, H.-Y. et al. Membrane curvature underlies actin reorganization in response to nanoscale surface topography.
_Proc. Natl Acad. Sci. USA_ 116, 23143–23151 (2019). Article ADS PubMed CAS PubMed Central Google Scholar * Fujiwara, T. K. et al. Confined diffusion of transmembrane proteins and
lipids induced by the same actin meshwork lining the plasma membrane. _Mol. Biol. Cell_ 27, 1101–1119 (2016). Article PubMed CAS PubMed Central Google Scholar * Li, J., Yan, J. &
Springer, T. A. Low-affinity integrin states have faster ligand-binding kinetics than the high-affinity state. _eLife_ 10, e73359 (2021). Article PubMed CAS PubMed Central Google Scholar
* Lemke, S. B., Weidemann, T., Cost, A.-L., Grashoff, C. & Schnorrer, F. A small proportion of Talin molecules transmit forces at developing muscle attachments in vivo. _PloS Biol._
17, e3000057 (2019). Article PubMed PubMed Central Google Scholar * Aretz, J., Aziz, M., Strohmeyer, N., Sattler, M. & Fässler, R. Talin and kindlin use integrin tail allostery and
direct binding to activate integrins. _Nat. Struct. Mol. Biol._ 30, 1913–1924 (2023). Article PubMed CAS PubMed Central Google Scholar * Liu, J. et al. Talin determines the nanoscale
architecture of focal adhesions. _Proc. Natl Acad. Sci. USA_ 112, E4864–E4873 (2015). Article PubMed CAS PubMed Central Google Scholar * Klapholz, B. et al. Alternative mechanisms for
talin to mediate integrin function. _Curr. Biol._ 25, 847–857 (2015). Article PubMed CAS PubMed Central Google Scholar * Swaminathan, V. et al. Actin retrograde flow actively aligns and
orients ligand-engaged integrins in focal adhesions. _Proc. Natl Acad. Sci. USA_ 114, 10648–10653 (2017). Article ADS PubMed CAS PubMed Central Google Scholar * Bataillé, L., Delon,
I., Ponte, J., Brown, N. H. & Jagla, K. Downstream of identity genes: muscle-type-specific regulation of the fusion process. _Dev. Cell_ 19, 317–328 (2010). Article PubMed Google
Scholar * Green, H. J., Griffiths, A. G., Ylänne, J. & Brown, N. H. Novel functions for integrin-associated proteins revealed by analysis of myofibril attachment in Drosophila. _eLife_
7, e35783 (2018). Article PubMed PubMed Central Google Scholar * Röper, K., Mao, Y. & Brown, N. H. Contribution of sequence variation in Drosophila actins to their incorporation into
actin-based structures in vivo. _J. Cell Sci._ 118, 3937–3948 (2005). Article PubMed Google Scholar * Friedl, K. et al. Assessing crosstalk in simultaneous multicolor single-molecule
localization microscopy. _Cell Rep. Methods_ 3, 100571 (2023). Article PubMed CAS PubMed Central Google Scholar * Butler, C. et al. Multi-dimensional spectral single molecule
localization microscopy. _Front. Bioinform._ 2, 813494 (2022). Article PubMed PubMed Central Google Scholar * Garcia, M. et al. Two-tiered coupling between flowing actin and immobilized
N-cadherin/catenin complexes in neuronal growth cones. _Proc. Natl Acad. Sci. USA_ 112, 6997–7002 (2015). Article ADS PubMed CAS PubMed Central Google Scholar * Teo, J. L., Lim, C.,
Yap, A. S. & Saw, T. A biologist’s guide to traction force microscopy using polydimethylsiloxane substrate for two-dimensional cell cultures. _STAR Protoc._ 1, 100098 (2020). Download
references ACKNOWLEDGEMENTS We thank Christel Poujol and Magali Mondin from the Bordeaux Imaging Center (BIC), the microscopy facilities of Cambridge Advance Imaging Centre (CAIC), Instituto
de Biomedicina de Sevilla (IBiS), and Centro Andaluz de Biología del Desarrollo (CABD) for technical assistance, the University of Cambridge Department of Genetics Fly Facility for embryo
injections. We would like to thank Frank Schnorrer for helpful discussions, Sven Bogdan for gift of fly lines and Benoît Ladoux for helpful discussions and gift of reagents, as well as and
Luis María Escudero for his technical support. We acknowledge financial support from the French Ministry of Research and CNRS, the Human Frontiers Science programme (Grant RGP0009/2017), and
the Fondation pour la Recherche Médicale (EQU202303016303). T.C. also received supported from the French government in the framework of the University of Bordeaux’s IdEx “Investments for
the Future” program/GPR LIGHT. C.H.F.E. also received support from the University of Cambridge Career Support Fund and María Zambrano contract funded by European Union—NextGenerationEU.
O.R., S.L.F., and G.G. are supported by the France BioImaging national infrastructure ANR-10-INBS-04. S.L.F. acknowledges the funding from the European Union’s Horizon 2020 research and
innovation program under grant agreement No. 871124 Laserlab-Europe (JRA ALTIS), and from ANR (MSM project ANR 17-CE09-0040-02). AUTHOR INFORMATION Author notes * These authors contributed
equally: Tianchi Chen, Cecilia H. Fernández-Espartero. * These authors jointly supervised this work: Nicholas H. Brown, Grégory Giannone. AUTHORS AND AFFILIATIONS * Interdisciplinary
Institute for Neuroscience, Université Bordeaux, CNRS, UMR 5297, Bordeaux, France Tianchi Chen, Mélanie Fabre, Olivier Rossier & Grégory Giannone * Department of Physiology, Development
and Neuroscience, University of Cambridge, Cambridge, UK Cecilia H. Fernández-Espartero, Benjamin Klapholz & Nicholas H. Brown * Instituto de Biomedicina de Sevilla, IBiS/Hospital
Universitario Virgen del Rocío/CSIC/Universidad de Sevilla and Departamento de Biología Celular, Universidad de Sevilla, Sevilla, Spain Cecilia H. Fernández-Espartero * Institut des sciences
Moléculaires d’Orsay, Université Paris Saclay, CNRS, UMR8214, Orsay, France Abigail Illand, Pierre Jouchet & Sandrine Lévêque-Fort * Department of Chemistry and Stanford Wu-Tsai
Neuroscience Institute, Stanford University, Stanford, CA, USA Ching-Ting Tsai, Yang Yang & Bianxiao Cui Authors * Tianchi Chen View author publications You can also search for this
author inPubMed Google Scholar * Cecilia H. Fernández-Espartero View author publications You can also search for this author inPubMed Google Scholar * Abigail Illand View author publications
You can also search for this author inPubMed Google Scholar * Ching-Ting Tsai View author publications You can also search for this author inPubMed Google Scholar * Yang Yang View author
publications You can also search for this author inPubMed Google Scholar * Benjamin Klapholz View author publications You can also search for this author inPubMed Google Scholar * Pierre
Jouchet View author publications You can also search for this author inPubMed Google Scholar * Mélanie Fabre View author publications You can also search for this author inPubMed Google
Scholar * Olivier Rossier View author publications You can also search for this author inPubMed Google Scholar * Bianxiao Cui View author publications You can also search for this author
inPubMed Google Scholar * Sandrine Lévêque-Fort View author publications You can also search for this author inPubMed Google Scholar * Nicholas H. Brown View author publications You can also
search for this author inPubMed Google Scholar * Grégory Giannone View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS T.C., C.H.F.E., N.H.B.,
and G.G. conceptualized the project and designed the experiments. T.C. performed embryo dissection, sptPALM, and super-resolution imaging experiments, developed MATLAB and Python code, and
performed data analysis. C.H.F.E. and B.K. conducted fly genetics and generated transgenic fly lines. C.H.F.E. performed SIM and Lightning imaging on fixed embryos. A.I., P.J., and S.L.F.
developed and performed 3D ModLoc super-resolution imaging experiments. C.-T.T., Y.Y., and B.C. designed and fabricated quartz nanopatterns. M.F. helped with fly husbandry and dissection.
O.R. and B.K. performed pioneering experiments of sptPALM. T.C., C.H.F.E, N.H.B., and G.G. wrote the manuscript. All authors discussed the results and commented on the manuscript.
CORRESPONDING AUTHORS Correspondence to Tianchi Chen, Nicholas H. Brown or Grégory Giannone. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW
PEER REVIEW INFORMATION _Nature Communications_ thanks Pakorn Kanchanawong, Anne Reversat and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer
review file is available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE DESCRIPTION OF ADDITIONAL SUPPLEMENTARY INFORMATION SUPPLEMENTARY MOVIE 1 SUPPLEMENTARY MOVIE 2 SUPPLEMENTARY MOVIE 3
SUPPLEMENTARY MOVIE 4 SUPPLEMENTARY MOVIE 5 SUPPLEMENTARY MOVIE 6 SUPPLEMENTARY MOVIE 7 SUPPLEMENTARY MOVIE 8 SUPPLEMENTARY MOVIE 9 SUPPLEMENTARY MOVIE 10 SUPPLEMENTARY MOVIE 11
SUPPLEMENTARY MOVIE 12 SUPPLEMENTARY MOVIE 13 SUPPLEMENTARY MOVIE 14 REPORTING SUMMARY SOURCE DATA SOURCE DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative
Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as
you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have
permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s
Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not
permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Chen, T., Fernández-Espartero, C.H., Illand, A. _et al._ Actin-driven
nanotopography promotes stable integrin adhesion formation in developing tissue. _Nat Commun_ 15, 8691 (2024). https://doi.org/10.1038/s41467-024-52899-x Download citation * Received: 17
November 2023 * Accepted: 24 September 2024 * Published: 07 October 2024 * DOI: https://doi.org/10.1038/s41467-024-52899-x SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative
