Cryo-em investigation of ryanodine receptor type 3
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Ryanodine Receptor isoform 3 (RyR3) is a large ion channel found in the endoplasmic reticulum membrane of many different cell types. Within the hippocampal region of the brain, it
is found in dendritic spines and regulates synaptic plasticity. It controls myogenic tone in arteries and is upregulated in skeletal muscle in early development. RyR3 has a unique functional
profile with a very high sensitivity to activating ligands, enabling high gain in Ca2+-induced Ca2+ release. Here we solve high-resolution cryo-EM structures of RyR3 in non-activating and
activating conditions, revealing structural transitions that occur during channel opening. Addition of activating ligands yields only open channels, indicating an intrinsically high open
probability under these conditions. RyR3 has reduced binding affinity to the auxiliary protein FKBP12.6 due to several sequence variations in the binding interface. We map disease-associated
sequence variants and binding sites for known pharmacological agents. The N-terminal region contains ligand binding sites for a putative chloride anion and ATP, both of which are targeted
by sequence variants linked to epileptic encephalopathy. SIMILAR CONTENT BEING VIEWED BY OTHERS PATHOLOGICAL CONFORMATIONS OF DISEASE MUTANT RYANODINE RECEPTORS REVEALED BY CRYO-EM Article
Open access 05 February 2021 STRUCTURAL BASIS FOR RYANODINE RECEPTOR TYPE 2 LEAK IN HEART FAILURE AND ARRHYTHMOGENIC DISORDERS Article Open access 15 September 2024 DUAL ROLE OF THE S5
SEGMENT IN TYPE 1 RYANODINE RECEPTOR CHANNEL GATING Article Open access 18 September 2024 INTRODUCTION Ryanodine receptors (RyRs) are ion channels that release Ca2+ from the endoplasmic (ER)
and sarcoplasmic reticulum (SR)1. With molecular weights >2 MDa, they are the largest ion channels currently known. Of the three isoforms (RyR1-3) that are found in mammals, the type 3
ryanodine receptor was first identified in mammalian brain2, but has since been found in many different cell types3. Within the brain, RyR3 is primarily found in the hippocampus and striatum
in adults4,5,6. Although RyR1 and RyR2 are also found throughout the central nervous system, RyR3 plays distinct roles4,7. For example, in primary hippocampal neurons, RyR3 is the only
isoform found in dendritic spines8. Activated by increases in cytosolic Ca2+, RyR3 likely amplifies Ca2+ signals originating from the plasma membrane at postsynaptic dendritic spines, which
then propagate to the dendrite and activate RyR2, which is more prevalent in this region. Accordingly, RyR3 has been shown to play a role in learning and memory, as its knock-down or sheer
knock-out leads to impaired spatial learning and neuronal plasticity9,10, reduced social interactions and hyperactivity11. Its role in the brain is underscored by several disorders. RyR3 is
overexpressed in hippocampal neurons of Alzheimer’s disease mouse models12,13, and its overexpression has also been associated with depression-like behavior14. Outside of the brain, RyR3 is
found in multiple tissues with specifically assigned roles in tracheal epithelium15, pre-adipocytes16, cerebral arteries17, but also in stomach, spleen, intestines, esophagus, kidneys and
more3. Given this diverse array of cell types, single nucleotide polymorphisms in RyR3 have been associated with a range of traits and disorders, including plasma adiponectin levels18,
atherosclerosis19,20,21, hypertension and diabetes22, Alzheimer’s22, fetal akinesia23, and neuroleptic malignant syndrome24. However, a direct causative link between RyR3 mutations and
disease has not been proven to date. Concomitant with a specialized role for RyR3, its intrinsic functional properties differ substantially from RyR1 and RyR2. Being the shortest of the
three isoforms, it has a higher sensitivity to activating ligands like Ca2+ and caffeine, to oxidating conditions, and is characterized by a much higher maximum open probability25,26,27,28.
This profile enables a specialized role in tissues where other isoforms are also expressed. In skeletal muscle, for example, RyR1 is the predominant isoform expressed in the SR membrane.
This is easily understood, as RyR1 is mechanically coupled to L-type Ca2+ channels in the T-tubule membrane, whereas RyR3 is not, and this effect is due to specific sequence differences in
distinct regions29,30,31. However, neonatal muscle lacks well-developed T-tubule systems, and are more reliant on Ca2+-induced Ca2+ release (CICR), which is strongly amplified by RyR3 by
virtue of its enhanced Ca2+ sensitivity32. The relative content of RyR3 in adults also varies substantially depending on the exact muscle, with much higher concentrations in diaphragm33,34.
This is particularly the case in extraocular muscle, where RyR3 is abundant, and its deletion strongly affects the muscle mechanical properties and vision35. In accordance with its role in
skeletal muscle, RyR3 sequence variants have been associated with nemaline myopathy36. High-resolution insights into RyR3 structure have been limited, with a crystal structure available for
the individual Repeat3&4 domain37, and unpublished structures of the Repeat1&2 domain (PDB entries 6UHA, 6UHB, 6UHE, 6UHH). Previous cryo-EM studies on full-length RyR3 have been
limited to very low resolutions ~30–40 Å38,39, thus not allowing any atomic-level interpretation. Here we present high-resolution cryo-EM structures of recombinantly expressed mammalian
RyR3. Our structural studies reveal a channel with a high intrinsic open probability, a distinct profile for binding small molecule ligands and altered interfaces for binding auxiliary
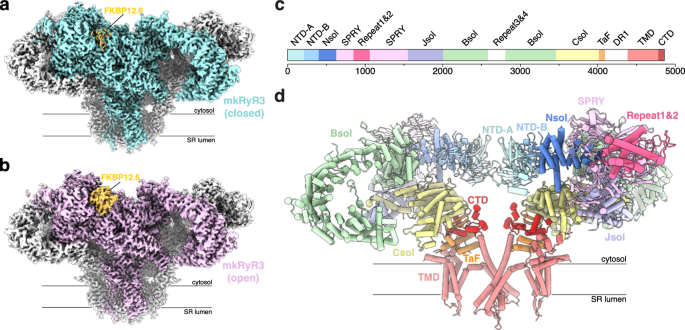
proteins like FKBP12.6. RESULTS STRUCTURES OF RYR3 IN NON-ACTIVATING AND ACTIVATING CONDITIONS We purified mink RyR3, recombinantly expressed in stable HEK293 cells, and determined cryo-EM
structures in two conditions: non-activating (5 mM EGTA) and activating (30 µM free Ca2+, 5 mM ATP, and 5 mM caffeine). These led to reconstructions at 2.89 and 3.22 Å global resolution,
respectively (Fig. 1; Supplementary Fig. 1 and 2; Supplementary Table 1). In both cases, FKBP12.6 was utilized to purify the channel, and was therefore present in the final reconstructions.
To determine the state of the pore in both conditions (open versus closed), we used classification on a masked transmembrane region and found that the non-activating condition gave rise to a
homogeneous population of particles with only closed pores. Conversely, the activating conditions only yielded open channels. This is in contrast with RyR1 and RyR2, where similar
activating conditions typically yield a mixture of open and closed channels1,40,41. This indicates an increased sensitivity of RyR3 for activating conditions and is in line with functional
data, which have shown a higher open probability of RyR3 compared to the other isoforms25,26,27,28. Using symmetry expansion and masked refinement, we highly improved the local resolution
for individual regions, yielding local resolutions in the pore domain of 2.6 and 2.8 Å for non-activating and activating conditions, respectively. The poorest resolution was obtained for the
Repeat1&2 domain (~6 Å) and the Repeat3&4 domain (nearly invisible), indicating high relative flexibility for these domains (Fig. 1a, b; Supplementary Figs. 1 and 2). An overview of
the various domains is shown in Fig. 1c, d. CONFORMATIONAL CHANGES IN CYTOSOLIC DOMAINS UPON CHANNEL OPENING Upon channel opening, the large cytosolic cap of RyR3 undergoes large outward
and downward motions away from the central fourfold symmetry axis (Fig. 2a; Supplementary Movie 1). These involve concerted movements in the N-terminal, Nsol, SPRY, Jsol, Bsol, Csol, and
C-terminal domains. Consequently, modulatory ligands or proteins that induce conformational changes in any of these cytosolic domains will likely also affect channel gating. Between the
closed and open states, the Bsol domain shows the largest downward movements (~10 Å) at the periphery (Fig. 2b). Of note, a pocket within the Bsol contains additional density not observed in
previous high-resolution structures of RyR142 or RyR241. We attribute this density to a loop further downstream in the Bsol sequence. The contacts are mediated by Arg3384 within the loop
and by Trp2513, Cys2517, His2872, and Asn2871 in the pocket. Closer to the fourfold symmetry axis, the N-terminal domains (NTD-A and NTD-B) from all four subunits form a near continuous
ring, with NTD-A of one subunit being juxtaposed to NTD-B′ of a neighboring subunit (Fig. 2c–f). Direct interactions may exist between Lys157 and Gln158 of NTD-A and Asp231′ and His230′ of
NTD-B′, but the side chain density for Lys157 and Gln158 is poorly resolved, and thus such interactions are, at most, transient. In the open state, however, the gap between NTD-A and NTD-B′
across subunits widens substantially, with a relative movement of ~4.5 Å measured by the increase in Cα-Cα distances between Lys157 and His230′. In contrast, the relative positions of NTD-A
and NTD-B within a subunit appear identical, in agreement with the more extensive interactions found between these domains. Immediately downstream of the last transmembrane region (S6) is a
C-terminal domain (CTD) containing a Zn2+-finger motif. In the closed state, the CTDs from all four subunits form a continuous ring with van der Waals interactions between Ala4846 and
Gly4847 on one subunit, and residues 4792′–4794′ on a neighboring subunit (Fig. 2g). In the open state of the channel, these interactions are lost, with a substantial gap between the CTDs
(Fig. 2h). These represent relative movements of 6 Å as measured by the Cα-Cα distances between Ala4846 and Pro4794′ of a neighboring subunit. Breaking these inter-subunit interactions
likely provides an energetic barrier for channel opening. CONFORMATIONAL CHANGES IN PORE-FORMING DOMAIN UPON CHANNEL OPENING The pore-forming domain is formed by the S5 and S6 helices of all
four subunits (Fig. 3a, b). The ion-conduction pathway is lined with many conserved residues that likely facilitate Ca2+ coordination and permeation. In the closed state, the hydrophobic
gate is formed by Ile4759 from all four subunits, resulting in a pore radius of less than 1 Å, thereby inhibiting Ca2+ conduction (Fig. 3c). In the open state, the narrowest points are
formed by Gly4716 and Gln4755, but with a minimum pore radius of ~3 Å that permits ion conduction. Upon channel opening, the S5 and S6 helices move outward, resulting in an increased
distance from 10.6 Å to 18.2 Å as measured by the Cα distance between Ile4759 of diagonally opposed subunits (Fig. 3d, e; Supplementary Movie 2). BINDING SITES FOR CA2+, CAFFEINE, AND ATP In
the RyR3 map obtained under activating conditions, we observed an unambiguous density for Ca2+, at an interface between two domains that are not adjacent in sequence: the central solenoid
(Csol) and C-terminal domain (CTD) (Fig. 4a, b). The Ca2+ ion is coordinated by the side chains of Glu3732 and Glu3806 in the Csol, and the Thr4823 carbonyl oxygen in the CTD. Binding of
Ca2+ to this region causes a relative decrease in the distance between these two domains (Supplementary Movie 3). This causes a pivoting motion of the CTD, which is then translated to the
associated S6 helices, thereby affecting channel gating. We also observed excellent density for caffeine, whose binding site is formed by 4 different regions: a cytosolic extension of the
loop connecting transmembrane helices S2 and S3, the CTD, the thumb-and-forefinger (TaF) domains, and a helix extending from the Csol. Binding of caffeine is obstructed by Trp4539, whose
side chain swings out to make place (Fig. 4c; Supplementary Movie 4). The extended S2-S3 loop moves away by 2.5 Å relative to the CTD to make space, whereas the Csol helix containing Phe3598
moves closer by ~8 Å to participate in caffeine binding. Excellent density is visible for ATP adjacent to the CTD, the extended S6 transmembrane helix, and the TaF domain (Fig. 4d). Several
structural changes can be perceived in the binding pocket comparing open and closed RyR3 (Fig. 5; Supplementary Movie 5). In closed RyR3, Arg4054, located in the TaF domain, is part of a
salt bridge network that involves Glu4051 (also in the TaF domain), which in turn links to Lys4643 in the S4-S5 linker (Fig. 5a). In the open RyR3, the S4-S5 linker has shifted, and Lys4643
is no longer involved in the salt bridge network. Arg4054 is now involved in ionic interactions with the ATP γ-phosphate, whereas Glu4051 now interacts with another Lys residue (Fig. 5b).
When one also takes intersubunit contacts into account, in the closed state of the channel the ATP β-phosphate group would be close (~3.5 Å) to Glu4066′ of a neighboring subunit, likely
providing some electrostatic repulsion (Fig. 5c). In the open state, Glu4066 is much further away, relieving the repulsion (Fig. 5d). We note that these changes around ATP are not due to ATP
alone but are observed when comparing open and closed channels. Indeed, previous structures of other RyR isoforms have shown that addition of ATP alone yields mostly closed channels40,43.
However, different electrostatic interactions that involve ATP do help explain why this ligand helps favor the open state when other activating ligands are also present40. LIGAND BINDING IN
THE NTD The N-terminal region is built up by the NTD-A and NTD-B domains, and the N-terminal solenoid (Nsol) (Fig. 1c, d). We observed a density compatible with a chloride anion, at the
interface where these three domains meet (Fig. 6a, b). This putative chloride is stabilized by four different arginine residues. Without it, one would expect significant electrostatic
repulsion due to these arginine residues, likely resulting in relative movements of these three domains that may alter channel gating. In RyR1, one of the arginines is replaced by a
histidine (Fig. 6c; Supplementary Fig. 3). This likely explains the absence of chloride binding in both crystal structures and cryo-EM structures of RyR140,44,45. RyR2 does contain four
arginines at this interface (Fig. 6d; Supplementary Fig. 3), but a corresponding chloride ion has not been assigned in any RyR2 cryo-EM structure to date. One crystal structure of the
isolated RyR2 N-terminal region has unambiguously shown the presence of a chloride anion in this area46. Closer inspection of available cryo-EM maps of RyR2 does show density compatible with
a chloride ion, suggesting that this is a feature that distinguishes RyR2 and RyR3 from RyR1. Unexpectedly, we observed density for another ATP molecule at the interface between the NTD-A
and NTD-B domains (Fig. 6a, e). The adenine ring fits in a mostly hydrophobic pocket, but with an additional hydrogen bond mediated by Arg285 in the NTD-B domain. The β and γ phosphate
groups are engaged in H-bond interactions, with an additional ionic interaction between the γ phosphate and Lys35 in NTD-A. This density has not been previously observed or reported in
cryo-EM structures of RyR1 or RyR2 determined in presence of ATP (Fig. 6f, g). The ability of RyR3 to bind ATP in this region is likely a unique feature of RyR3. His205 involved in
coordinating β phosphate of ATP is mutated to an asparagine and alanine in RyR1 and RyR2, respectively (Supplementary Fig. 3). Additional differences between RyR3 and RyR2 include
substitution of RyR3 Ala56 by Ser in RyR2, likely introducing steric hindrance. RyR3 Ser272, which interacts with the β phosphate, is replaced by Ala in RyR2. Comparing the binding pocket
for ATP in the open and closed RyR3, there are only subtle differences in sidechain conformations, suggesting that either state is compatible with ATP binding. Its binding at the NTD-A:NTD-B
interface may confer structural rigidity. This ATP molecule is 81 Å away from the one bound to CTD (Supplementary Fig. 4). Whereas direct coupling between ATP binding at both sites is thus
unlikely, it is possible that they influence one another’s binding through long-range allosteric effects. In open-state structures of RyR1 and RyR2, the local resolution of the NTD-A and
NTD-B domains is frequently lower, indicating higher relative mobility. Binding of both chloride and ATP may prevent this, contributing to the high local resolution of this area in open RyR3
(Supplementary Fig. 2) and also indirectly affecting channel gating. Indeed, post-translational modifications and mutations at the interfaces between the N-terminal domains can
allosterically affect channel opening47, via a mechanism that links these to changes at the NTD-A:NTD-B′ across subunits, or to altered interactions between the NTDs and the central solenoid
(Csol)45,46,48. Thus, the rigidity induced by binding of two ligands in this area may also have effects on channel gating. ISOFORM-SPECIFIC DIFFERENCES RyR3 has been found to have a higher
Po compared to RyR1 and RyR2 in various conditions25,26,27,28. In addition to the chloride and ATP binding sites in the N-terminal region, we note several other differences between RyR3 and
the other isoforms. FKBP12 and FKBP12.6 (FK506-binding proteins of 12 and 12.6 kDa, respectively) associate with RyRs, stabilizing their closed state49. It was previously noted that RyR3 has
a lowered affinity for these proteins50. We used a GST-FKBP12.6 fusion protein to purify RyR3, indicating that it still retains binding. However, we observed weaker density for FKBP12.6 in
our closed RyR3 structure (Fig. 1a), suggesting a reduced occupancy or higher relative mobility. In contrast, the FKBP12.6 density in the open RyR3 was better defined. A careful comparison
of the FKBP12.6:RyR3 and FKBP12.6:RyR1 interfaces shows that there are various distinct differences that explain a lower affinity (Fig. 7a, b). In RyR1, His1300 in the SPRY3 domain interacts
with Gln32 in FKBP12.6, but in RyR3 this is replaced by Ser1299, which no longer interacts. Lys36 in FKBP12.6 hydrogen bonds with Asn636 in the RyR1 SPRY1 domain, but this has been replaced
by the bulkier Arg634 in RyR3, which cannot form a hydrogen bond due to steric hindrance and electrostatic repulsion. Val91 in FKBP12.6 forms van der Waals interactions with Ser1687 in the
RyR1 junctional solenoid (Jsol) but has been replaced with Gly1582 in RyR3. These differences likely underlie the reduced affinity of FKBP12.6 for RyR3. The cryo-EM density in the Bsol
domain, particularly in the later portion, appears to be more resolved compared to existing RyR2 structures41,51 (Supplementary Fig. 5a, b). Sequence alignment in this region reveals a
12-residue insertion in RyR2 that is absent in RyR1 and RyR3 near the end of the Bsol domain (Supplementary Fig. 5c). This insertion may contribute to the longer linker connecting the Bsol
and Csol domains in RyR2. As a result, the Bsol domain in RyR2 is less constrained, possibly leading to higher conformational heterogeneity and lower visibility. Several cryo-EM structures
of RyR1 have revealed an additional transmembrane segment that precedes the S1 helix in sequence40,52,53. This helix makes contacts with helices S1 and S4 of one subunit, and S5′ of another
subunit (Fig. 7c, d). We did not observe density for the S0 helix in our RyR3 structures, although the proposed sequence is largely conserved. In RyR1, Val4339 in S0 is juxtaposed with
Val4820 at the end of S4, but both of these are replaced by Phe in RyR3, which would create a steric clash. Therefore, the S0 helix is likely more flexible in RyR3. DISEASE-ASSOCIATED
VARIANTS To date, more than 1200 RyR3 sequence variants, found in patients with different disorders, have been reported in the ClinVar database54. Around 75% of these have been associated
with epileptic encephalopathy, an epileptic condition that leads to cerebral dysfunction55,56. Supplementary Table 3 summarizes RyR3 mutations, reported in peer-reviewed literature and found
in patients with a wide range of disorders. Mapping these onto the RyR3 structure (Fig. 8a) shows that most of these are found in the SPRY domains and in the Bsol region. A total of 10 of
these mutations affect residues buried within the structures and are thus more likely to change function (Supplementary Table 3). For residues that are solvent exposed, a change in function
is less likely. We investigated sequence variants at the ligand binding sites. So far, no variants reported in peer-reviewed literature are at these binding sites, but 7 variants, linked to
epileptic encephalopathy in ClinVar database, are found in the binding sites for ATP and chloride within the N-terminal region (Supplementary Table 4; Fig. 8b, c). Of note, mutation of
arginine residues coordinating the chloride ion in RyR2 have been firmly linked to catecholaminergic polymorphic ventricular tachycardia (CPVT)47,57. For example, functional experiments on
the RYR2 R420Q and R420W mutation, equivalent to RyR3 R412W, have indicated a gain-of-function phenotype58,59. Knock-in mice and hiPSC-derived cardiomyocytes with the RyR2 R420Q mutation
recapitulate the CPVT phenotype and show structural alterations59. Crystallographic investigation of these mutations in the N-terminal disease hot spot have shown a change in the relative
domain orientations46. Thus, it is very likely that the equivalent R412W mutation in RyR3 also introduces a gain-of-function and may be causative for epileptic encephalopathy. Knock-in mouse
studies will be needed to investigate this further. DISCUSSION We sought to elucidate the structure of RyR3, and solved cryo-EM structures in different experimental conditions.
Surprisingly, we found that adding a cocktail of activating ligands (caffeine, ATP, and 30 µM free Ca2+) led to a homogeneous population of only open channels, in stark contrast with studies
with the RyR1 where a mixture of open and closed channels is observed under such conditions40. This indicates an intrinsically higher open probability for RyR3 than for RyR1. This matches
previous functional experiments that have indicated an increased sensitivity of RyR3 to activating ligands and a higher maximum open probability25,26,27,28, but we note that cryo-EM studies
of RyR2 in the presence of activating ligands have also exclusively shown open channels51,60. What might be the cause for this increased open probability? The main sequence differences
between the three RyR isoforms are located in the divergent regions (DR1–DR3), large areas that are intrinsically disordered1. Of note, DR2 is contained within a flexible loop of the SPRY3
domain and is almost non-existent for RyR3 (Supplementary Data 1). Although these regions likely impart distinct functional changes, they have not been visualized in any cryo-EM structure of
RyRs to date, and thus may have little impact on the proportion of open isolated RyRs observed in the cryo-EM conditions. However, the DRs likely confer further isoform-specific difference
in physiological context, where many additional binding partners are present. Several differences in the RyR3 structure may explain its unique functional properties, including several
sequence changes that are predicted to lower the affinity for FKBP12 and FKBP12.6. The latter proteins are known to bind RyR1 and RyR2 with high affinity and are thought to stabilize the
closed state49,61. Despite the sequence changes in the binding site, RyR3 clearly has not lost its ability to associate with FKBPs, since the initial purification step included GST-FBKP12.6
as a bait protein (see methods). We did notice a decreased occupancy for FKBP12.6 in our closed RyR3 structure, suggesting that some was lost during the subsequent purification step. It is
unclear why this was more pronounced for the closed state structure but is in agreement with previous observations that RyR3 binds FKBPs with lower affinity50. We also found the ligand
profile to differ. RyR3 retains the previously identified binding sites for caffeine, ATP and activating Ca2+, but an additional ATP binding site is found at the interface between the NTD-A
and NTD-B domains, where it may confer rigidity and stabilize the relative domain orientation. This site is likely unique for RyR3 as it has not been observed before for RyR1 or RyR2, and
there are differences in the residues that make up the binding pocket. We also identified a putative chloride anion at the interface between the NTD-A, NTD-B and Nsol domains. It is
coordinated by arginine residues, all of which are conserved in RyR2, but not RyR1. Previous experiments with RyR2 have shown that mutation of a single arginine is sufficient to obliterate
chloride binding to this site46 and has been linked to CPVT58. The relationship between RyRs and disease has been firmly established for both RyR1 and RyR2. Through many functional
experiments and knock-in mouse models, it is very clear that many sequence variants cause a gain- or loss-of-function phenotype, causing CPVT or cardiomyopathy (RyR2) or malignant
hyperthermia, central core disease, or other myopathies (RyR1)47,62. A plethora of sequence variants have been identified for RyR3, including hundreds that have been found in patients with
epileptic encephalopathy. Although the large number suggests a causative link, this remains to be proven. However, comparing known causative mutations in other isoforms suggests that at
least a subset of the RyR3 variants may alter function, and thus also cause disease. For example, the R420Q and R420W mutations in RyR2 cause a gain-of-function phenotype and are causative
of CPVT. They both affect the binding of chloride to the RyR2 N-terminal region and cause a relative reorientation of the N-terminal domains46. An equivalent mutation in RyR3 (R412W) has
been found in a patient with epileptic encephalopathy and is thus likely to have similar structural and functional consequences. Of note, many single nucleotide polymorphisms in RyR3 are
located in intronic regions. They are less likely to directly affect the function of the individual RyR3 but may have an impact on the total amount of RyR3 protein. Since RyR3 inherently has
a much higher open probability, disturbing the balance between RyR3 and the other isoforms thus presents another mechanism for an overall gain-of-function. Whether RyR3 mutations truly
cause disease remains to be demonstrated, and thus it will be of interest to generate animal models of several different RyR3 sequence variants to test for the role of RyR3 in disease.
METHODS EXPRESSION AND PURIFICATION OF HIS-GST-FKBP12.6 We used a human FKBP12.6 (2–108) with an N-terminal His6-GST-TEV-tag52. The construct was transformed into _E. coli_ strain Rosetta
(DE3). Cells were grown in auto-induction media63 at 37 °C to an optical density of 1, at which the temperature was changed to 20 °C and left shaking overnight. The cell pellet was obtained
by centrifuging at 5000 × _g_ for 20 min. The pellet was re-suspended in lysis buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, 5 mM BME) supplemented with 10 μg/mL DNAse I, 1 mM MgCl2,
0.1 mg/mL lysozyme, 0.5 mM PMSF, and 5 mM imidazole. Cells were lysed by sonication. Cellular debris was removed by centrifugation at 44,000 × _g_ for 45 min at 4 °C. The supernatant was
loaded onto Ni-NTA resin (Qiagen) by batch binding, and incubated with gentle shaking for 30 min at 4 °C. The resin was washed with lysis buffer containing 30 mM imidazole and eluted with
lysis buffer containing 500 mM imidazole. The protein was dialyzed overnight in buffer A (10 mM Tris pH 8.8, 10 mM NaCl, 5 mM BME), and applied onto an HiLoad 16/10 Q Sepharose High
Performance column (Cytiva) equilibrated with buffer A and eluted using a linear gradient of buffer B (10 mM Tris pH 8.8, 1 M NaCl, 5 mM BME). The protein was further purified by size
exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column (Cytiva) in FPLC buffer (20 mM HEPES pH 7.5, 250 mM NaCl, 1 mM TCEP). The final purified protein was concentrated to around
10 mg/mL (measured by NanoDrop), flash-frozen in liquid nitrogen, and stored at −70 °C for later use. EXPRESSION OF RYR3 We used HEK293T cells stably expressing mink RyR327. The cells were
cultured in 150-mm dishes with Minimum Essential Medium α (MEM α; Gibco Cat #12561056) supplemented with 10% heat-inactivated Fetal Bovine Serum (Gibco, Cat #12484028), 10,000 U/mL
penicillin-streptomycin solution, and additional 2 mM L-glutamine at 37 °C with 5% CO2. Cells were maintained in culture below passage 10 and positively selected with 800 μg/mL G418 if kept
in continued culture for more than a month. Upon confluency, plates were placed on ice, and cells were washed with cold Dulbecco’s Phosphate Buffered Saline (DPBS), harvested with a scraper,
and centrifuged at 250 × _g_ for 5 minutes at 4 °C. Centrifugation cycles were repeated until all cells from 50−100 plates were collected. The cell pellet was flash-frozen in liquid
nitrogen and stored at −70 °C for later use. PURIFICATION OF RYR3 All steps were performed at 4 °C. Cell pellets from 100 dishes were thawed and re-suspended in buffer A (20 mM Tris-maleate
pH 6.8, 75 mM NaCl, 10% sucrose, 1 mM DTT, 1:1000 of protease inhibitor cocktail (PIC; Millipore Sigma Cat# 539134)) and lysed by sonication. Cellular debris was removed by centrifugation at
4400 × _g_ for 10 min. The supernatant was ultracentrifuged at 100,000 × _g_ for 60 min. The membrane pellet was collected, flash-frozen in liquid nitrogen, and stored at −70 °C for later
use. The membrane fraction after thawing was solubilized in buffer B (25 mM HEPES pH 7.5, 500 mM NaCl, 2 mM DTT, 1% GDN, 1:1000 of PIC for 1 h. The solubilized mixture was ultracentrifuged
at 100,000 × _g_ for 60 min. The supernatant was mixed with 5 mg of His-GST-FKBP12.6 and incubated for 1 h. Next, 1.5 mL of Glutathione Sepharose 4B resin (Cytiva) was added for 1 h. The
resin was poured into a gravity column, washed with buffer C (25 mM HEPES pH 7.5, 200 mM NaCl, 2 mM DTT, 0.02% GDN, 1:1000 of PIC) and eluted with buffer D (75 mM HEPES pH 8, 200 mM NaCl, 2
mM DTT, 0.02% GDN, 1:1000 of PIC, 15 mM glutathione). The His-GST tag was removed by overnight incubation with 150 µg of Tobacco Etch Virus (TEV) protease. The mixture was incubated with 0.2
mL of TALON resin (Cytiva) for 30 min to capture His-GST and His-TEV. The gravity column flow-through was applied onto a 1-mL HiTrap Heparin HP column (Cytiva) equilibrated in buffer E (20
mM HEPES pH 7.5, 50 mM NaCl, 2 mM DTT, 0.02% GDN, 1:1000 of PIC, 5 mM EGTA) and eluted with Buffer E containing 500 mM NaCl. For the activating condition, buffer E contains a final free Ca2+
concentration of 30 µM, using a total of 2 mM EGTA, 5 mM ATP, and 5 mM caffeine instead of 5 mM EGTA. The total concentration of Ca2+ to obtain 30 µM free Ca2+ was calculated with Ca-EGTA
Calculator TS v1.364 and the final free Ca2+ concentration was verified using a PerfectIon Combination Calcium Electrode unit (Mettler Toledo). The elution is concentrated to 100 µL then
diluted with 150 µL of Buffer E, making final [NaCl] = ~250 mM. The elution is then concentrated to ~5 mg/mL (measured with Nanodrop), flash-frozen in liquid nitrogen, and stored at −70 °C.
ELECTRON MICROSCOPY Separate grids were prepared under non-activating and activating conditions, respectively. Holey gold grids (UltrAuFoil Au 300 mesh, R 1.2/1.3) were glow-discharged for 2
min. 2.5 µL of RyR3-FKBP12.6 sample (non-activating or activating condition) was applied, blotted for 3 s (blot force of 7 to 10) using ashless blotting paper (Whatman) and subsequently
plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) at 4 °C and 100% humidity. Cryo-EM grids were screened on a Glacios electron microscope operating at 200 kV
and equipped with a Falcon 3 camera (Thermo Fisher Scientific). High-resolution data were collected with a Titan Krios G2 electron microscope operating at 300 kV and equipped with a Falcon
4i camera and a Selectris energy filter (Thermo Fisher Scientific). Microscope operations and data collection were carried out using EPU software (Thermo Fisher Scientific). Movies were
gathered in super-resolution counting mode at a calibrated magnification of 130,000×, corresponding to 0.96 Å per physical pixel. For the non-activating dataset, a total dose of 50 e−/Å2
using a dose rate of 10.20 e−/pixel/s was delivered to 1404 frames in EER format with a defocus range of −1 to −2 µm. For the activating dataset, a total dose of 50 e−/Å2 using a dose rate
of 12.06 e−/pixel/s was delivered to 1188 frames in EER format with a defocus range of −0.5 to −2 µm. CRYOEM DATA PROCESSING Detailed schematics of the cryo-EM data processing pipeline for
both non-activating and activating datasets are summarized in Supplementary Figs. 1 and 2, respectively. All steps were performed in cryoSPARC65 (v. 4.3–4.4.1) unless otherwise indicated.
Movies were patch motion-corrected and curated based on ice thickness, defocus values, contrast transfer function (CTF) resolution estimation (lower than 6 Å were kept), full-frame motion,
astigmatism, and average intensity. Particle picking was carried out using both crYOLO66 (v. 1.9.7) and template picker in cryoSPARC. Templates used in template picker were generated from 2D
classification of crYOLO-picked particles. After two rounds of 2D classification, particles from both pipelines were combined, and duplicates were removed. The particles were further
cleaned using iterative rounds of ab-initio reconstruction and heterogeneous refinement. A consensus refinement was performed using non-uniform refinement. Masked 3D classification was
performed using a TMD mask (residues 4016–4859) to classify the various pore conformations. Particles with the same pore state were combined, processed with reference-based motion
correction, and refined with non-uniform refinement with C4 symmetry imposed. Pixel calibration was performed by comparing real-space correlation with a crystal structure of the N-terminal
domains of RyR1 (PDB: 2XOA). Local refinements were performed to improve local resolution using four separate masks. The first mask consists of the N-terminal, SPRY, and Repeat1&2
domains (residues 1–1616) + FKBP12.6. The second mask consists of the Jsol, Csol, and Bsol domains (residues 1617–2492, 3487–4015). The third mask contains the Bsol and Repeat3&4 domain
(residues 2170–3462). The fourth mask contains the TMD (residues 4016–4859). The first three local refinements used C4-symmetry expanded particles while the fourth refinement used
non-symmetry expanded particles with C4 symmetry imposed. The resulting locally refined maps were combined in ChimeraX67 to generate a composite map. Fourier shell correlation (FSC) plots,
orientation distributions and refinement statistics are presented in Supplementary Figs. 1 and 2 and Supplementary Table 1. Mask-corrected FSC curves were calculated based on the FSC = 0.143
criterion. The local resolution estimations for the entire channel and each of the locally refined maps were calculated using the FSC = 0.5 criterion. Cryo-EM density visualization was done
in UCSF ChimeraX67. MODEL BUILDING AND REFINEMENT A model of human RyR3 predicted by AlphaFold68 was manually docked into the composite map using ChimeraX67. The mismatched residues were
manually changed to that of mink RyR3 in Coot69, and the model was improved through iterative cycles of manual building in Coot69 and real-space refinement in PHENIX70. For the Repeat1&2
domain, which showed weak cryo-EM density, crystal structures of human RyR3 Repeat1&2 domain (PDB: 6UHB & 6UHA) were rigid-body-docked into the composite maps of non-activating and
activating datasets in ChimeraX67, and mismatched residues were manually changed to that of mink RyR3 in Coot69. Structural model validation was performed using PHENIX71 and MolProbity72.
Validation statistics are summarized in Supplementary Table 1. Structural images were prepared with UCSF ChimeraX67. Quantification of the pore radius in Fig. 3 was calculated using HOLE73.
HOMOLOGY MODELING OF HUMAN RYR3 The homology model of human RyR3 was constructed with MODELER74 (v. 10.5) using mink RyR3 structure as the template. The sequence alignment was performed
using Clustal Omega75. REPORTING SUMMARY Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY Atomic
coordinates of RyR3 have been deposited in the Protein Data Bank (PDB) with the following accession codes: 9C1E (non-activating) and 9C1F (activating). The composite, consensus, and four
local refinement maps of closed state RyR3 (non-activating) have been deposited in the Electron Microscopy Data Bank (EMDB) with the following accession codes: EMDB-45116
[https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45116], EMDB-45035 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45035], EMDB-45107 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45107], EMDB-45108
[https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45108], EMDB-45109 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45109], and EMDB-45110 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45110]. The
composite, consensus, and four local refinement maps of open state RyR3 (activating) have been deposited in the EMDB with the following accession codes: EMDB-45117
[https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45117], EMDB-45111 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45111], EMDB-45112 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45112], EMDB-45113
[https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45113], EMDB-45114 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45114], and EMDB-45115 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-45115]. The
following previously published PDB codes were used for comparison: 2XOA, 6UHA, 6UHB, 6UHE, 6UHH, 7TZC,7U9T, 5TAL, 7UA9, 8DVE. The source data underlying Supplementary Figs. 1a and 2a is
provided as a Source Data file. Source data are provided with this paper. REFERENCES * Woll, K. A. & Van Petegem, F. Calcium-release channels: structure and function of IP(3) receptors
and ryanodine receptors. _Physiol. Rev._ 102, 209–268 (2022). Article CAS PubMed Google Scholar * Hakamata, Y., Nakai, J., Takeshima, H. & Imoto, K. Primary structure and
distribution of a novel ryanodine receptor/calcium release channel from rabbit brain. _FEBS Lett._ 312, 229–235 (1992). Article CAS PubMed Google Scholar * Lanner, J. T., Georgiou, D.
K., Joshi, A. D. & Hamilton, S. L. Ryanodine receptors: structure, expression, molecular details, and function in calcium release. _Cold Spring Harb. Perspect. Biol._ 2, a003996 (2010).
Article CAS PubMed PubMed Central Google Scholar * Torres, R. & Hidalgo, C. Subcellular localization and transcriptional regulation of brain ryanodine receptors. Functional
implications. _Cell Calcium_ 116, 102821 (2023). Article CAS PubMed Google Scholar * Mori, F., Fukaya, M., Abe, H., Wakabayashi, K. & Watanabe, M. Developmental changes in expression
of the three ryanodine receptor mRNAs in the mouse brain. _Neurosci. Lett._ 285, 57–60 (2000). Article CAS PubMed Google Scholar * Murayama, T. & Ogawa, Y. Properties of Ryr3
ryanodine receptor isoform in mammalian brain. _J. Biol. Chem._ 271, 5079–5084 (1996). Article CAS PubMed Google Scholar * Hidalgo, C. & Paula-Lima, A. RyR-mediated calcium release
in hippocampal health and disease. _Trends Mol. Med_ 30, 25–36 (2024). Article CAS PubMed Google Scholar * Vega-Vasquez, I. et al. Hippocampal dendritic spines express the RyR3 but not
the RyR2 ryanodine receptor isoform. _Biochem Biophys. Res Commun._ 633, 96–103 (2022). Article CAS PubMed Google Scholar * Balschun, D. et al. Deletion of the ryanodine receptor type 3
(RyR3) impairs forms of synaptic plasticity and spatial learning. _EMBO J._ 18, 5264–5273 (1999). Article CAS PubMed PubMed Central Google Scholar * Galeotti, N. et al. Different
involvement of type 1, 2, and 3 ryanodine receptors in memory processes. _Learn Mem._ 15, 315–323 (2008). Article CAS PubMed PubMed Central Google Scholar * Matsuo, N. et al.
Comprehensive behavioral phenotyping of ryanodine receptor type 3 (RyR3) knockout mice: decreased social contact duration in two social interaction tests. _Front, Behav. Neurosci._ 3, 3
(2009). Article PubMed Google Scholar * Supnet, C., Grant, J., Kong, H., Westaway, D. & Mayne, M. Amyloid-beta-(1-42) increases ryanodine receptor-3 expression and function in neurons
of TgCRND8 mice. _J. Biol. Chem._ 281, 38440–38447 (2006). Article CAS PubMed Google Scholar * Supnet, C., Noonan, C., Richard, K., Bradley, J. & Mayne, M. Up-regulation of the type
3 ryanodine receptor is neuroprotective in the TgCRND8 mouse model of Alzheimer’s disease. _J. Neurochem._ 112, 356–365 (2010). Article CAS PubMed Google Scholar * Nakamura-Maruyama, E.
et al. Ryanodine receptors are involved in the improvement of depression-like behaviors through electroconvulsive shock in stressed mice. _Brain Stimul._ 14, 36–47 (2021). Article PubMed
Google Scholar * Schmidt, G. et al. A fixed 20:1 combination of cafedrine/theodrenaline increases cytosolic Ca(2+) concentration in human tracheal epithelial cells via ryanodine
receptor-mediated Ca(2+) release. _Sci. Rep._ 13, 16216 (2023). Article ADS CAS PubMed PubMed Central Google Scholar * Tsai, S. H. et al. Knockdown of RyR3 enhances adiponectin
expression through an atf3-dependent pathway. _Endocrinology_ 154, 1117–1129 (2013). Article CAS PubMed Google Scholar * Lohn, M. et al. Regulation of calcium sparks and spontaneous
transient outward currents by RyR3 in arterial vascular smooth muscle cells. _Circ. Res._ 89, 1051–1057 (2001). Article CAS PubMed Google Scholar * Chang, Y. C. et al. Genome-wide
linkage analysis and regional fine mapping identified variants in the RYR3 gene as a novel quantitative trait locus for circulating adiponectin in Chinese population. _Medicine_ 95, e5174
(2016). Article CAS PubMed PubMed Central Google Scholar * Shendre, A. et al. RYR3 gene variants in subclinical atherosclerosis among HIV-infected women in the Women’s Interagency HIV
Study (WIHS). _Atherosclerosis_ 233, 666–672 (2014). Article CAS PubMed PubMed Central Google Scholar * Shrestha, S. et al. Replication of RYR3 gene polymorphism association with cIMT
among HIV-infected whites. _AIDS_ 26, 1571–1573 (2012). Article CAS PubMed Google Scholar * Zhao, C. et al. Association of the RYR3 gene polymorphisms with atherosclerosis in elderly
Japanese population. _BMC Cardiovasc. Disord._ 14, 6 (2014). Article PubMed PubMed Central Google Scholar * Gong, S. et al. Polymorphisms within RYR3 gene are associated with risk and
age at onset of hypertension, diabetes, and Alzheimer’s disease. _Am. J. Hypertens._ 31, 818–826 (2018). Article CAS PubMed Google Scholar * Pergande, M. et al. The genomic and clinical
landscape of fetal akinesia. _Genet. Med._ 22, 511–523 (2020). Article CAS PubMed Google Scholar * Dettling, M., Sander, T., Weber, M. & Steinlein, O. K. Mutation analysis of the
ryanodine receptor gene isoform 3 (RYR3) in recurrent neuroleptic malignant syndrome. _J. Clin. Psychopharmacol._ 24, 471–473 (2004). Article PubMed Google Scholar * Murayama, T. et al.
Further characterization of the type 3 ryanodine receptor (RyR3) purified from rabbit diaphragm. _J. Biol. Chem._ 274, 17297–17308 (1999). Article CAS PubMed Google Scholar * Perez, C.
F., Lopez, J. R. & Allen, P. D. Expression levels of RyR1 and RyR3 control resting free Ca2+ in skeletal muscle. _Am. J. Physiol. Cell Physiol._ 288, C640–C649 (2005). Article CAS
PubMed Google Scholar * Rossi, D. et al. RyR1 and RyR3 isoforms provide distinct intracellular Ca2+ signals in HEK 293 cells. _J. Cell Sci._ 115, 2497–2504 (2002). Article CAS PubMed
Google Scholar * Murayama, T. & Ogawa, Y. Characterization of type 3 ryanodine receptor (RyR3) of sarcoplasmic reticulum from rabbit skeletal muscles. _J. Biol. Chem._ 272, 24030–24037
(1997). Article CAS PubMed Google Scholar * Protasi, F. et al. RYR1 and RYR3 have different roles in the assembly of calcium release units of skeletal muscle. _Biophys. J._ 79, 2494–2508
(2000). Article ADS CAS PubMed PubMed Central Google Scholar * Perez, C. F., Mukherjee, S. & Allen, P. D. Amino acids 1-1,680 of ryanodine receptor type 1 hold critical
determinants of skeletal type for excitation-contraction coupling. Role of divergence domain D2. _J. Biol. Chem._ 278, 39644–39652 (2003). Article CAS PubMed Google Scholar * Perez, C.
F., Voss, A., Pessah, I. N. & Allen, P. D. RyR1/RyR3 chimeras reveal that multiple domains of RyR1 are involved in skeletal-type E-C coupling. _Biophys. J._ 84, 2655–2663 (2003). Article
ADS CAS PubMed PubMed Central Google Scholar * Yang, D. et al. RyR3 amplifies RyR1-mediated Ca(2+)-induced Ca(2+) release in neonatal mammalian skeletal muscle. _J. Biol. Chem._ 276,
40210–40214 (2001). Article CAS PubMed Google Scholar * Conti, A., Gorza, L. & Sorrentino, V. Differential distribution of ryanodine receptor type 3 (RyR3) gene product in mammalian
skeletal muscles. _Biochem. J._ 316, 19–23 (1996). Article CAS PubMed PubMed Central Google Scholar * Conti, A., Reggiani, C. & Sorrentino, V. Selective expression of the type 3
isoform of ryanodine receptor Ca2+ release channel (RyR3) in a subset of slow fibers in diaphragm and cephalic muscles of adult rabbits. _Biochem. Biophys. Res. Commun._ 337, 195–200 (2005).
Article CAS PubMed Google Scholar * Eckhardt, J. et al. Extraocular muscle function is impaired in ryr3 (-/-) mice. _J. Gen. Physiol._ 151, 929–943 (2019). Article CAS PubMed PubMed
Central Google Scholar * Nilipour, Y. et al. Ryanodine receptor type 3 (RYR3) as a novel gene associated with a myopathy with nemaline bodies. _Eur. J. Neurol._ 25, 841–847 (2018). Article
CAS PubMed Google Scholar * Yuchi, Z., Lau, K. & Van Petegem, F. Disease mutations in the ryanodine receptor central region: crystal structures of a phosphorylation hot spot domain.
_Structure_ 20, 1201–1211 (2012). Article CAS PubMed Google Scholar * Sharma, M. R., Jeyakumar, L. H., Fleischer, S. & Wagenknecht, T. Three-dimensional structure of ryanodine
receptor isoform three in two conformational states as visualized by cryo-electron microscopy. _J. Biol. Chem._ 275, 9485–9491 (2000). Article CAS PubMed Google Scholar * Liu, Z. et al.
Three-dimensional reconstruction of the recombinant type 3 ryanodine receptor and localization of its amino terminus. _Proc. Natl. Acad. Sci. USA_ 98, 6104–6109 (2001). Article ADS CAS
PubMed PubMed Central Google Scholar * des Georges, A. et al. Structural basis for gating and activation of RyR1. _Cell_ 167, 145–157.e17 (2016). Article CAS PubMed PubMed Central
Google Scholar * Miotto, M. C. et al. Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment. _Sci. Adv._ 8, eabo1272
(2022). Article CAS PubMed PubMed Central Google Scholar * Melville, Z. et al. A drug and ATP binding site in type 1 ryanodine receptor. _Structure_ (2022). * Cholak, S. et al.
Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives. _Structure_ (2023). * Tung, C. C., Lobo, P. A., Kimlicka, L. & Van Petegem, F. The amino-terminal disease
hotspot of ryanodine receptors forms a cytoplasmic vestibule. _Nature_ 468, 585–588 (2010). Article ADS CAS PubMed Google Scholar * Kimlicka, L., Lau, K., Tung, C. C. & Van Petegem,
F. Disease mutations in the ryanodine receptor N-terminal region couple to a mobile intersubunit interface. _Nat. Commun._ 4, 1506 (2013). Article ADS PubMed Google Scholar * Kimlicka,
L. et al. The cardiac ryanodine receptor N-terminal region contains an anion binding site that is targeted by disease mutations. _Structure_ 21, 1440–1449 (2013). Article CAS PubMed
Google Scholar * Pancaroglu, R. & Van Petegem, F. Calcium channelopathies: structural insights into disorders of the muscle excitation-contraction complex. _Annu. Rev. Genet_ 52,
373–396 (2018). Article CAS PubMed Google Scholar * van den Bersselaar, L. R. et al. RYR1 variant c.38T>G, p.Leu13Arg causes hypersensitivity of the ryanodine receptor-1 and is
pathogenic for malignant hyperthermia. _Br. J. Anaesth._ 127, e63–e65 (2021). Article PubMed Google Scholar * Van Petegem, F. Ryanodine receptors: structure and function. _J. Biol. Chem._
287, 31624–31632 (2012). Article PubMed PubMed Central Google Scholar * Fessenden, J. D. et al. Divergent functional properties of ryanodine receptor types 1 and 3 expressed in a
myogenic cell line. _Biophys. J._ 79, 2509–2525 (2000). Article ADS CAS PubMed PubMed Central Google Scholar * Gong, D. et al. Modulation of cardiac ryanodine receptor 2 by calmodulin.
_Nature_ 572, 347–351 (2019). Article ADS CAS PubMed Google Scholar * Ma, R. et al. Structural basis for diamide modulation of ryanodine receptor. _Nat. Chem. Biol._ 16, 1246–1254
(2020). Article CAS PubMed Google Scholar * Haji-Ghassemi, O. et al. Cryo-EM analysis of scorpion toxin binding to ryanodine receptors reveals subconductance that is abolished by PKA
phosphorylation. _Sci. Adv._ 9, eadf4936 (2023). Article CAS PubMed PubMed Central Google Scholar * Landrum, M. J. et al. ClinVar: improvements to accessing data. _Nucleic Acids Res._
48, D835–D844 (2020). Article CAS PubMed Google Scholar * Khan, S. & Al Baradie, R. Epileptic encephalopathies: an overview. _Epilepsy Res. Treat._ 2012, 403592 (2012). PubMed
PubMed Central Google Scholar * Kalser, J. & Cross, J. H. The epileptic encephalopathy jungle—from Dr West to the concepts of aetiology-related and developmental encephalopathies.
_Curr. Opin. Neurol._ 31, 216–222 (2018). Article PubMed Google Scholar * Medeiros-Domingo, A. et al. The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed
previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome: a comprehensive open reading frame mutational analysis.
_J. Am. Coll. Cardiol._ 54, 2065–2074 (2009). Article CAS PubMed PubMed Central Google Scholar * Tang, Y., Tian, X., Wang, R., Fill, M. & Chen, S. R. Abnormal termination of Ca2+
release is a common defect of RyR2 mutations associated with cardiomyopathies. _Circ. Res._ 110, 968–977 (2012). Article CAS PubMed PubMed Central Google Scholar * Yin, L. et al.
Impaired binding to junctophilin-2 and nanostructural alteration in CPVT mutation. _Circ. Res._ 129, e35–e52 (2021). Article CAS PubMed PubMed Central Google Scholar * Chi, X. et al.
Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators. _Proc. Natl. Acad. Sci. USA_ 116, 25575–25582 (2019). Article ADS CAS PubMed
PubMed Central Google Scholar * Brillantes, A. B. et al. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. _Cell_ 77, 513–523 (1994). Article
CAS PubMed Google Scholar * Betzenhauser, M. J. & Marks, A. R. Ryanodine receptor channelopathies. _Pflug. Arch._ 460, 467–480 (2010). Article CAS Google Scholar * Studier, F. W.
Protein production by auto-induction in high density shaking cultures. _Protein Expr. Purif._ 41, 207–234 (2005). Article CAS PubMed Google Scholar * Schoenmakers, T. J., Visser, G. J.,
Flik, G. & Theuvenet, A. P. CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. _Biotechniques_ 12, 876–879 (1992). 870-4. Google Scholar *
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. _Nat. Methods_ 14, 290–296 (2017). Article
CAS PubMed Google Scholar * Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. _Commun. Biol._ 2, 218 (2019). Article PubMed PubMed
Central Google Scholar * Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. _Protein Sci._ 30, 70–82 (2021). Article CAS PubMed
Google Scholar * Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. _Nature_ 596, 583–589 (2021). Article ADS CAS PubMed PubMed Central Google Scholar *
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. _Acta Crystallogr. D Biol. Crystallogr_ 66, 486–501 (2010). Article ADS CAS PubMed PubMed Central
Google Scholar * Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. _Acta Crystallogr. D Struct. Biol._ 74, 531–544 (2018). Article ADS CAS PubMed
PubMed Central Google Scholar * Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. _Acta Crystallogr. D Struct. Biol._ 74, 814–840 (2018).
Article ADS CAS PubMed PubMed Central Google Scholar * Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. _Protein Sci._ 27,
293–315 (2018). Article CAS PubMed Google Scholar * Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions
of ion channel structural models. _J. Mol. Graph_ 14, 376 (1996). 354-60. Article CAS PubMed Google Scholar * Webb, B. & Sali, A. Comparative protein structure modeling using
MODELLER. _Curr. Protoc. Bioinform._ 54, 5 6 1–5 6 37 (2016). Article Google Scholar * Madeira, F. et al. Search and sequence analysis tools services from EMBL-EBI in 2022. _Nucleic Acids
Res._ 50, W276–W279 (2022). Article CAS PubMed PubMed Central Google Scholar * Persson, I. Hydrated metal ions in aqueous solution: How regular are their structures? _Pure Appl. Chem._
82, 1901–1917 (2010). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work is funded by Canadian Institutes of Health Research grant PJT-159601 (F.V.P.), Fonds de
Recherche du Québec–Santé fellowship BF7-310936 (Y.S.C.), and Michael Smith Health Research BC Research Trainee award RT-2023-3133 (Y.S.C.). Cryo-EM grids were prepared and collected at the
High Resolution Macromolecular Electron Microscopy (HRMEM) facility at the University of British Columbia (https://cryoem.med.ubc.ca). We thank Claire Atkinson, Joeseph Felt, Liam Worrall
and Natalie Strynadka. HRMEM is funded by the Canadian Foundation of Innovation and the British Columbia Knowledge Development Fund. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department
of Biochemistry and Molecular Biology, the Life Sciences Centre, University of British Columbia, Vancouver, BC, Canada Yu Seby Chen, Maricela Garcia-Castañeda, Maria Charalambous & Filip
Van Petegem * Department of Molecular and Developmental Medicine, University of Siena, Siena, Italy Daniela Rossi & Vincenzo Sorrentino Authors * Yu Seby Chen View author publications
You can also search for this author inPubMed Google Scholar * Maricela Garcia-Castañeda View author publications You can also search for this author inPubMed Google Scholar * Maria
Charalambous View author publications You can also search for this author inPubMed Google Scholar * Daniela Rossi View author publications You can also search for this author inPubMed Google
Scholar * Vincenzo Sorrentino View author publications You can also search for this author inPubMed Google Scholar * Filip Van Petegem View author publications You can also search for this
author inPubMed Google Scholar CONTRIBUTIONS F.V.P. and V.S. conceived the project. M.C. performed mammalian protein expression with guidance by D.R. for maintaining the stable cell line.
Y.S.C. & M.G. prepared cryo-EM samples. Y.S.C. performed cryo-EM data processing, model building and refinement. Y.S.C. and F.V.P. analyzed the structures and wrote the first version of
the manuscript, with edits provided by all other authors. CORRESPONDING AUTHOR Correspondence to Filip Van Petegem. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Communications_ thanks Deshun Gong and Manjuli Sharma for their contribution to the peer review of this work. A peer review file is
available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY MOVIE 1 SUPPLEMENTARY MOVIE 2 SUPPLEMENTARY MOVIE 3 SUPPLEMENTARY MOVIE 4
SUPPLEMENTARY MOVIE 5 SUPPLEMENTARY DATA 1 REPORTING SUMMARY SOURCE DATA SOURCE DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give
appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission
under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons
licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by
statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Chen, Y.S., Garcia-Castañeda, M., Charalambous, M. _et al._ Cryo-EM
investigation of ryanodine receptor type 3. _Nat Commun_ 15, 8630 (2024). https://doi.org/10.1038/s41467-024-52998-9 Download citation * Received: 30 May 2024 * Accepted: 27 September 2024 *
Published: 05 October 2024 * DOI: https://doi.org/10.1038/s41467-024-52998-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable
link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
