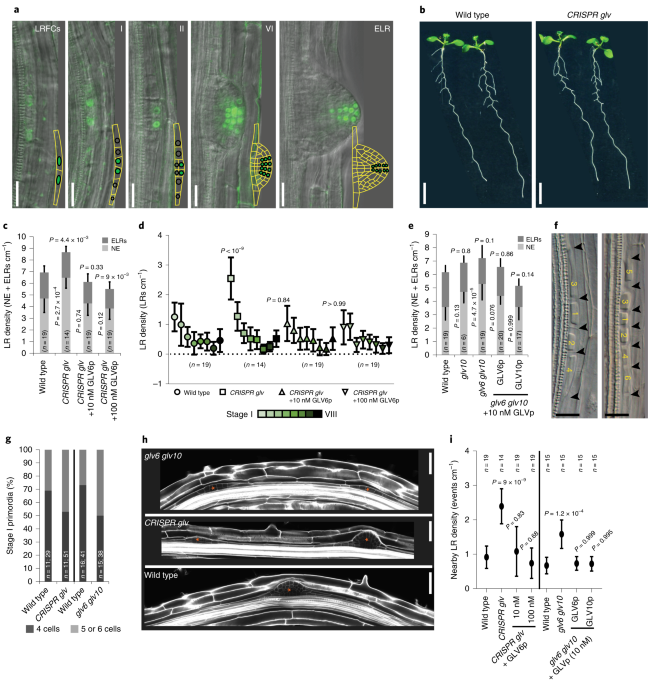
Golven peptide signalling through rgi receptors and mpk6 restricts asymmetric cell division during lateral root initiation
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT During lateral root initiation, lateral root founder cells undergo asymmetric cell divisions that generate daughter cells with different sizes and fates, a prerequisite for correct
primordium organogenesis. An excess of the GLV6/RGF8 peptide disrupts these initial asymmetric cell divisions, resulting in more symmetric divisions and the failure to achieve lateral root
organogenesis. Here, we show that loss-of-function _GLV6_ and its homologue _GLV10_ increase asymmetric cell divisions during lateral root initiation, and we identified three members of the
RGF1 INSENSITIVE/RGF1 receptor subfamily as likely GLV receptors in this process. Through a suppressor screen, we found that MITOGEN-ACTIVATED PROTEIN KINASE6 is a downstream regulator of
the GLV pathway. Our data indicate that GLV6 and GLV10 act as inhibitors of asymmetric cell divisions and signal through RGF1 INSENSITIVE receptors and MITOGEN-ACTIVATED PROTEIN KINASE6 to
restrict the number of initial asymmetric cell divisions that take place during lateral root initiation. Access through your institution Buy or subscribe This is a preview of subscription
content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access
subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more
Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS:
* Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS DISTINCT MECHANISMS ORCHESTRATE THE CONTRA-POLARITY OF
IRK AND KOIN, TWO LRR-RECEPTOR-KINASES CONTROLLING ROOT CELL DIVISION Article Open access 11 January 2022 ANTAGONISTIC CLE PEPTIDE PATHWAYS SHAPE ROOT MERISTEM TISSUE PATTERNING Article 28
October 2024 SHR AND SCR COORDINATE ROOT PATTERNING AND GROWTH EARLY IN THE CELL CYCLE Article Open access 31 January 2024 DATA AVAILABILITY The data supporting the findings in this study
are available from the corresponding author upon reasonable request. REFERENCES * Moreno-Risueno, M. A. et al. Oscillating gene expression determines competence for periodic _Arabidopsis_
root branching. _Science_ 329, 1306–1311 (2010). CAS PubMed PubMed Central Google Scholar * Xuan, W. et al. Cyclic programmed cell death stimulates hormone signaling and root development
in _Arabidopsis_. _Science_ 351, 384–387 (2016). CAS PubMed Google Scholar * De Smet, I. et al. Receptor-like kinase ACR4 restricts formative cell divisions in the _Arabidopsis_ root.
_Science_ 322, 594–597 (2008). PubMed Google Scholar * De Smet, I. et al. Bimodular auxin response controls organogenesis in _Arabidopsis_. _Proc. Natl Acad. Sci. USA_ 107, 2705–2710
(2010). PubMed Google Scholar * De Rybel, B. et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. _Curr. Biol._ 20,
1697–1706 (2010). PubMed Google Scholar * Hofhuis, H. et al. Phyllotaxis and rhizotaxis in _Arabidopsis_ are modified by three PLETHORA transcription factors. _Curr. Biol._ 23, 956–962
(2013). CAS PubMed Google Scholar * Casimiro, I. et al. Auxin transport promotes _Arabidopsis_ lateral root initiation. _Plant Cell_ 13, 843–852 (2001). CAS PubMed PubMed Central
Google Scholar * Lucas, M. et al. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. _Proc. Natl Acad. Sci. USA_ 110, 5229–5234 (2013). CAS
PubMed Google Scholar * von Wangenheim, D. et al. Rules and self-organizing properties of post-embryonic plant organ cell division patterns. _Curr. Biol._ 26, 439–449 (2016). Google
Scholar * Meng, L., Buchanan, B. B., Feldman, L. J. & Luan, S. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in _Arabidopsis_. _Proc. Natl
Acad. Sci. USA_ 109, 1760–1765 (2012). CAS PubMed Google Scholar * Fernandez, A. et al. Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling
peptides reveals their role in lateral root and hair formation. _Plant Physiol._ 161, 954–970 (2013). CAS PubMed Google Scholar * Fernandez, A. et al. The GLV6/RGF8/CLEL2 peptide
regulates early pericycle divisions during lateral root initiation. _J. Exp. Bot._ 66, 5245–5256 (2015). CAS PubMed PubMed Central Google Scholar * Peterson, B. A. et al. Genome-wide
assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in _Arabidopsis_. _PLoS ONE_ 11, e0162169 (2016). PubMed PubMed Central Google Scholar * Komori,
R., Amano, Y., Ogawa-Ohnishi, M. & Matsubayashi, Y. Identification of tyrosylprotein sulfotransferase in _Arabidopsis_. _Proc. Natl Acad. Sci. USA_ 106, 15067–15072 (2009). CAS PubMed
Google Scholar * Zhou, W. et al. _Arabidopsis_ tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. _Plant
Cell_ 22, 3692–3709 (2010). CAS PubMed PubMed Central Google Scholar * Matsuzaki, Y., Ogawa-Ohnishi, M., Mori, A. & Matsubayashi, Y. Secreted peptide signals required for
maintenance of root stem cell niche in _Arabidopsis_. _Science_ 329, 1065–1067 (2010). CAS PubMed Google Scholar * Whitford, R. et al. GOLVEN secretory peptides regulate auxin carrier
turnover during plant gravitropic responses. _Dev. Cell_ 22, 678–685 (2012). CAS PubMed Google Scholar * Wu, T. et al. An _Arabidopsis thaliana_ copper-sensitive mutant suggests a role of
phytosulfokine in ethylene production. _J. Exp. Bot._ 66, 3657–3667 (2015). CAS PubMed PubMed Central Google Scholar * Lopez-Bucio, J. S. et al. _Arabidopsis thaliana_ mitogen-activated
protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. _J. Exp. Bot._ 65, 169–183 (2014). CAS PubMed Google Scholar * Xu, J. & Zhang,
S. Mitogen-activated protein kinase cascades in signaling plant growth and development. _Trends Plant Sci._ 20, 56–64 (2015). CAS PubMed Google Scholar * Ditengou, F. A. et al. Mechanical
induction of lateral root initiation in _Arabidopsis thaliana_. _Proc. Natl Acad. Sci. USA_ 105, 18818–18823 (2008). CAS PubMed Google Scholar * Laskowski, M. et al. Root system
architecture from coupling cell shape to auxin transport. _PLoS Biol._ 6, e307 (2008). PubMed PubMed Central Google Scholar * Ortiz-Morea, F. A. et al. Danger-associated peptide signaling
in _Arabidopsis_ requires clathrin. _Proc. Natl Acad. Sci. USA_ 113, 11028–11033 (2016). CAS PubMed Google Scholar * Song, W. et al. Signature motif-guided identification of receptors
for peptide hormones essential for root meristem growth. _Cell Res._ 26, 674–685 (2016). CAS PubMed PubMed Central Google Scholar * Ou, Y. et al. RGF1 INSENSITIVE 1 to 5, a group of LRR
receptor-like kinases, are essential for the perception of root meristem growth factor 1 in _Arabidopsis thaliana_. _Cell Res._ 26, 686–698 (2016). CAS PubMed PubMed Central Google
Scholar * Shinohara, H., Mori, A., Yasue, N., Sumida, K. & Matsubayashi, Y. Identification of three LRR-RKs involved in perception of root meristem growth factor in _Arabidopsis_.
_Proc. Natl Acad. Sci. USA_ 113, 3897–3902 (2016). CAS PubMed Google Scholar * Voss, U. et al. The circadian clock rephases during lateral root organ initiation in _Arabidopsis thaliana_.
_Nat. Commun._ 6, 7641 (2015). PubMed PubMed Central Google Scholar * Peret, B. et al. Auxin regulates aquaporin function to facilitate lateral root emergence. _Nat. Cell Biol._ 14,
991–998 (2012). CAS PubMed Google Scholar * Fukaki, H., Tameda, S., Masuda, H. & Tasaka, M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14
gene of _Arabidopsis_. _Plant J._ 29, 153–168 (2002). CAS PubMed Google Scholar * Okushima, Y. et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in
_Arabidopsis thaliana_: unique and overlapping functions of ARF7 and ARF19. _Plant Cell_ 17, 444–463 (2005). CAS PubMed PubMed Central Google Scholar * Benkova, E. et al. Local,
efflux-dependent auxin gradients as a common module for plant organ formation. _Cell_ 115, 591–602 (2003). CAS PubMed Google Scholar * Dubrovsky, J. G. et al. Auxin acts as a local
morphogenetic trigger to specify lateral root founder cells. _Proc. Natl Acad. Sci. USA_ 105, 8790–8794 (2008). CAS PubMed Google Scholar * Cho, S. K. et al. Regulation of floral organ
abscission in _Arabidopsis thaliana_. _Proc. Natl Acad. Sci. USA_ 105, 15629–15634 (2008). CAS PubMed Google Scholar * Jewaria, P. K. et al. Differential effects of the peptides Stomagen,
EPF1 and EPF2 on activation of MAP kinase MPK6 and the SPCH protein level. _Plant Cell Physiol._ 54, 1253–1262 (2013). CAS PubMed Google Scholar * Turing, A. M. The chemical basis of
morphogenesis. _Phil. Trans. R. Soc. Lond. B_ 237, 37–72 (1952). Google Scholar * Torii, K. U. Two-dimensional spatial patterning in developmental systems. _Trends Cell Biol._ 22, 438–446
(2012). PubMed Google Scholar * Toyokura, K. et al. Lateral inhibition by a peptide hormone–receptor cascade during _Arabidopsis_ lateral root founder cell formation. _Dev. Cell_ 48, 64–75
(2019). CAS PubMed Google Scholar * Kumpf, R. P. et al. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. _Proc. Natl
Acad. Sci. USA_ 110, 5235–5240 (2013). CAS PubMed Google Scholar * Zhu, Q. et al. A MAPK cascade downstream of IDA-HAE/HSL2 ligand–receptor pair in lateral root emergence. _Nat. Plants_
5, 414–423 (2019). CAS PubMed Google Scholar * Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. _Plant
Physiol._ 133, 462–469 (2003). CAS PubMed PubMed Central Google Scholar * Barbez, E. et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants.
_Nature_ 485, 119–122 (2012). CAS PubMed Google Scholar * Jun, J. et al. Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in _Arabidopsis_.
_Plant Physiol._ 154, 1721–1736 (2010). CAS PubMed PubMed Central Google Scholar * Schneeberger, K. et al. SHOREmap: simultaneous mapping and mutation identification by deep sequencing.
_Nat. Methods_ 6, 550–551 (2009). CAS PubMed Google Scholar * Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. _Nat. Methods_ 9,
671–675 (2012). CAS PubMed PubMed Central Google Scholar * Schindelin, J., Arganda-Carreras, I. & Frise, E. et al. Fiji: an open-source platform for biological-image analysis. _Nat.
Methods_ 9, 676–682 (2012). CAS PubMed Google Scholar * Malamy, J. E. & Benfey, P. N. Organization and cell differentiation in lateral roots of _Arabidopsis thaliana_. _Development_
124, 33–44 (1997). CAS PubMed Google Scholar * Xuan, W., Opdenacker, D., Vanneste, S. & Beeckman, T. Long-term in vivo imaging of luciferase-based reporter gene expression in
_Arabidopsis_ roots. _Methods Mol. Biol._ 1761, 177–190 (2018). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS This research was supported by FWO postdoctoral (A.F., grant
no. 1293817N) and doctoral (J.J., grant no. 1168218N) fellowships, a China Scholarship Council grant (K.X., no. 201606350134) and a National Science Foundation Plant Genome Research Program
Grant (Z.L.N., no. PGRP-1841917). We thank M. Njo for help with preparing the figures, V. Storme for guidance and assistance with the statistical analysis and D. Savatin for training with
the MPK6 phosphorylation experiments. AUTHOR INFORMATION Author notes * These authors contributed equally: Ana I. Fernandez, Nick Vangheluwe. AUTHORS AND AFFILIATIONS * Department of Plant
Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium Ana I. Fernandez, Nick Vangheluwe, Ke Xu, Joris Jourquin, Lucas Alves Neubus Claus, Stefania Morales-Herrera, Boris
Parizot, Hugues De Gernier, Qiaozhi Yu, Andrzej Drozdzecki, Davy Opdenacker, Eugenia Russinova & Tom Beeckman * VIB Center for Plant Systems Biology, Ghent, Belgium Ana I. Fernandez,
Nick Vangheluwe, Ke Xu, Joris Jourquin, Lucas Alves Neubus Claus, Stefania Morales-Herrera, Boris Parizot, Hugues De Gernier, Qiaozhi Yu, Andrzej Drozdzecki, Davy Opdenacker, Eugenia
Russinova & Tom Beeckman * Laboratory of Molecular Cell Biology, KU Leuven, Kasteelpark, Leuven, Belgium Stefania Morales-Herrera * VIB Center for Microbiology, Kasteelpark, Leuven,
Belgium Stefania Morales-Herrera * Department of Life Science and Biotechnology, Faculty of Life and Environmental Science, Shimane University, Matsue, Japan Takanori Maruta * Department of
Organic Chemistry and Macromolecular Chemistry, Ghent University, Ghent, Belgium Kurt Hoogewijs, Willem Vannecke & Annemieke Madder * Department of Biology, University of North Carolina,
Chapel Hill, NC, USA Brenda Peterson & Zachary L. Nimchuk Authors * Ana I. Fernandez View author publications You can also search for this author inPubMed Google Scholar * Nick
Vangheluwe View author publications You can also search for this author inPubMed Google Scholar * Ke Xu View author publications You can also search for this author inPubMed Google Scholar *
Joris Jourquin View author publications You can also search for this author inPubMed Google Scholar * Lucas Alves Neubus Claus View author publications You can also search for this author
inPubMed Google Scholar * Stefania Morales-Herrera View author publications You can also search for this author inPubMed Google Scholar * Boris Parizot View author publications You can also
search for this author inPubMed Google Scholar * Hugues De Gernier View author publications You can also search for this author inPubMed Google Scholar * Qiaozhi Yu View author publications
You can also search for this author inPubMed Google Scholar * Andrzej Drozdzecki View author publications You can also search for this author inPubMed Google Scholar * Takanori Maruta View
author publications You can also search for this author inPubMed Google Scholar * Kurt Hoogewijs View author publications You can also search for this author inPubMed Google Scholar * Willem
Vannecke View author publications You can also search for this author inPubMed Google Scholar * Brenda Peterson View author publications You can also search for this author inPubMed Google
Scholar * Davy Opdenacker View author publications You can also search for this author inPubMed Google Scholar * Annemieke Madder View author publications You can also search for this author
inPubMed Google Scholar * Zachary L. Nimchuk View author publications You can also search for this author inPubMed Google Scholar * Eugenia Russinova View author publications You can also
search for this author inPubMed Google Scholar * Tom Beeckman View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS A.I.F. designed the project.
A.I.F. generated the _iGLV6_ line, performed the EMS mutagenesis screen and analysed the EMS mutants’ identity with help from N.V., A.D., D.O. and T.M. A.I.F. and K.X. phenotypically
characterized the _CRISPR glv_ mutants, and generated and characterized the _rgi_ mutants and reporter lines. N.V. generated and characterized the _CRISPR glv6_ mutants. N.V., K.X., J.J. and
S.M.-H. phenotypically characterized the _mpk6_ mutants. J.J., S.M.-H. and H.D.G. characterized the cross-talk between the auxin and GLV pathways. Q.Y. generated the _RGI4_ reporter line.
B. Parizot performed the in silico expression analysis. L.A.N.C., N.V. and E.R. performed the MPK6 phosphorylation experiments. B. Peterson and Z.L.N. generated the _CRISPR glv_ mutants.
K.H., W.V. and A.M. synthesized the peptides. J.J. and A.I.F. performed the statistical analysis. A.I.F., N.V. and T.B. wrote the manuscript with input from all authors. T.B. provided
guidance and advice on the project, the experiments and the analysis of the results. CORRESPONDING AUTHOR Correspondence to Tom Beeckman. ETHICS DECLARATIONS COMPETING INTERESTS The authors
declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Plants_ thanks Melinka Butenko, Juan Xu and the other, anonymous, reviewer(s) for their contribution to
the peer review of this work. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED
DATA FIG. 1 GLV6 AND 10 ACT REDUNDANTLY DURING LR INITIATION. A–C, Phenotypic characterization of _CRISPR glv6_ mutants compared to wild type (8 dag, n = 12). Quantification of root length
(a), all primordium stages density (B) and non-emerged primordia (NE) and emerged (E) LR density (C). D, Quantification of root length in the _CRISPR glv_ mutant compared to wild type. E,
Quantification of all primordium stages density in the _glv6glv10_ mutant germinated on MS or on 10 nM of GLV6p/GLV10p (8 dag). Charts show mean values ± s.d. (B, E) or s.e.m (C).
Significant differences compared to wild type are shown and were determined using one-way ANOVA (A, D) or a GEE model (B-C, E). In E, only significant differences in stage I primordia are
displayed. For full statistical analysis see Supplementary Table 2. n.s.: no significant differences were found between mutants and wild type. F, Example of nearby primordia frequently found
in _glv_ mutants. The lower picture shows a higher magnification image of the framed area in the upper picture for each genotype. Scale bars represent 50 μm. EXTENDED DATA FIG. 2
SUPPRESSION OF THE _GLV6__OE_ PHENOTYPE AND LR DEFECTS IN _MPK6_ MUTANTS. A, Suppression of the _GLV6__OE_ phenotype in _mpk6_ mutants after LR initiation was induced by gravistimulation of
the primary root. This experiment was done three times with similar results. B, Quantification of all primordium stages in reported _mpk6_ mutants compared to wild type (8 dag). Chart
represents mean values ± s.d. A GEE model was used. n.s.: no significant differences were found between mutants and wild type. For full statistical analysis see Supplementary Table 2. Scale
bars represent 20 μm. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figs. 1–9, Table 3 and methods. REPORTING SUMMARY SUPPLEMENTARY TABLES 1 AND 2 Table 1. Amino acid
sequences predicted to be encoded in _CRISPR glv6_ or _glv_ mutants. The GLV6 pre-propeptide is shown as a reference. The presumed mature GLV6 peptide sequence is highlighted in bold, and
stop codons are depicted as asterisks. Amino acid sequences different from the corresponding GLV wild-type precursor are italicized in the mutants. Table 2. Statistical analysis of the
primordia stages and LR density in _glv_ and _mpk6_ mutants compared to controls. _P_ values indicating significant differences are highlighted in green. See Methods for details on the
statistical analysis. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Fernandez, A.I., Vangheluwe, N., Xu, K. _et al._ GOLVEN peptide signalling through
RGI receptors and MPK6 restricts asymmetric cell division during lateral root initiation. _Nat. Plants_ 6, 533–543 (2020). https://doi.org/10.1038/s41477-020-0645-z Download citation *
Received: 01 April 2019 * Accepted: 24 March 2020 * Published: 11 May 2020 * Issue Date: May 2020 * DOI: https://doi.org/10.1038/s41477-020-0645-z SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative
