Pik3ca and map3k1 alterations imply luminal a status and are associated with clinical benefit from pan-pi3k inhibitor buparlisib and letrozole in er+ metastatic breast cancer
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Clinical trials have demonstrated the efficacy of combining phosphoinositide 3-kinase (PI3K) inhibitors with endocrine therapies in hormone therapy-refractory breast cancer.
However, biomarkers of PI3K pathway dependence in ER+ breast cancer have not been fully established. Hotspot mutations in the alpha isoform of PI3K (_PIK3CA_) are frequent in ER+ disease and
may identify tumors that respond to PI3K inhibitors. It is unclear whether PIK3CA mutations are the only biomarker to suggest pathway dependence and response to therapy. We performed
correlative molecular characterization of primary and metastatic tissue from patients enrolled in a phase Ib study combining buparlisib (NVP-BKM-120), a pan-PI3K inhibitor, with letrozole in
ER+, human epidermal growth factor-2 (HER2)-negative, metastatic breast cancer. Activating mutations in _PIK3CA_ and inactivating _MAP3K1_ mutations marked tumors from patients with
clinical benefit (≥6 months of stable disease). Patients harboring mutations in both genes exhibited the greatest likelihood of clinical benefit. In ER+ breast cancer cell lines,
siRNA-mediated knockdown of MAP3K1 did not affect the response to buparlisib. In a subset of patients treated with buparlisib or the PI3Kα inhibitor alpelisib each with letrozole where PAM50
analysis was performed, nearly all tumors from patients with clinical benefit had a luminal A subtype. Mutations in MAP3K1 in ER+ breast cancer may be associated with clinical benefit from
combined inhibition of PI3K and ER, but we could not ascribe direct biological function therein, suggesting they may be a surrogate for luminal A status. We posit that luminal A tumors may
be a target population for this therapeutic combination. SIMILAR CONTENT BEING VIEWED BY OTHERS _PIK3CA_ MUTATIONS IN ENDOCRINE-RESISTANT BREAST CANCER Article Open access 31 May 2024 THE
INTERPLAY BETWEEN FOXO3 AND FOXM1 INFLUENCES SENSITIVITY TO AKT INHIBITION IN PIK3CA AND PIK3CA/PTEN ALTERED ESTROGEN RECEPTOR POSITIVE BREAST CANCER Article Open access 22 April 2025 PI3K
ACTIVATION PROMOTES RESISTANCE TO ERIBULIN IN HER2-NEGATIVE BREAST CANCER Article 15 March 2021 INTRODUCTION Estrogen receptor-positive (ER+) breast cancer is the most common clinical
subtype of breast cancer, and afflicts over 140,000 patients each year in the US.1 Although endocrine therapy (aromatase inhibition or selective estrogen receptor modulators/selective
estrogen receptor down regulators; SERMs/SERDs) is largely active against ER+ disease, many patients demonstrate or develop therapeutic resistance to endocrine therapy, which can occur
through a variety of clinically established mechanisms. Such mechanisms include amplification of _fibroblast growth factor receptor-1_ (_FGFR1_), _estrogen receptor-alpha_ (_ESR1_)
mutations, and aberrant activation of cyclin-dependent-kinase 4/6 (CDK4/6) and PI3K/mTOR. CDK4/6 inhibitors (ribociclib, palbociclib, abemaciclib) are now an approved standard of care in ER+
breast cancer, and their utilization has substantially improved patient outcome. mTOR inhibitors are also approved, but their impact has not been as substantial. Nonetheless, the mechanisms
of resistance to endocrine therapy in the clinic are still largely unknown in many of patients. _PIK3CA_, encoding the p110α subunit of PI3K, is recurrently mutated in 40–50% of ER+ tumors,
suggesting a dependency of ER+ breast cancer cells on this pathway.2,3,4,5 Of these, as many as 80% occur in hotspots within the helical and kinase domains of the p110α isozyme.2 It has
been well established that these alterations induce PI3K activity resulting in transformation when overexpressed in human mammary epithelial cells, and can induce mammary tumor formation in
transgenic mice.6,7 Given the role of PI3K in supporting proliferation, survival, and hormone receptor pathway activity, it is not surprising that activation of this pathway is hypothesized
to provide cellular avenues to circumvent endocrine therapy sensitivity. Unfortunately, single agent PI3K inhibitors have not yet demonstrated meaningful activity in clinical trials. Several
factors have contributed to their underwhelming performance. First, disruption of the PI3K pathway is followed by feedback activation of compensatory pathways that limit sustained drug
target inhibition and suppression of PIP3 production (reviewed in ref. 8). One of these adaptive mechanisms is stimulation of C-peptide and insulin secretion, which activates insulin
receptors in tumors, further limiting the suppression of PI3K.9 Second, therapeutic inhibition with pan-PI3K and PI3K inhibitors is associated with toxicities, such as hyperglycemia, rash,
and gastrointestinal side effects as a result of inhibition of wild type PI3K isozymes, hence also preventing sustained and specific inhibition of mutant PIK3CA. Third, mutations in the PI3K
pathway are associated with somatic alterations in parallel oncogenic pathways and receptor tyrosine kinases that also stimulate PI3K. Finally, not all clinical trials to date have
rigorously selected patients with tumors that are dependent on PI3K signaling and that, therefore, are likely to respond to PI3K antagonists. Early clinical trials suggest PIK3CA mutations
are a biomarker of such dependence10,11, but that may still not capture the whole repertoire of cancers likely to benefit from these drugs. Herein, we analyzed next generation sequencing
data from a targeted panel of known recurrently mutated genes in cancer in a cohort of patients from two phase Ib trials of buparlisib and alpelisib, each in combination with the aromatase
inhibitor letrozole. The goal of this analysis was to determine possible biomarkers aside from _PIK3CA_ mutations status that were associated with clinical benefit to the combination.
Patients in both trials were endocrine refractory (at least one line of prior failed endocrine therapy), representing an area of therapy that could be greatly impacted by new treatment
options. While patients demonstrating clinical benefit (defined as CR/PR or SD ≥ 6 months) were enriched for _PIK3CA_ mutations, these alterations were a suboptimal predictor of response.
However, the co-presence of alterations in _MAP3K1_, a putative tumor suppressor, with PIK3CA mutations was associated with a significant clinical benefit rate (CBR). This finding spurred
molecular analyses to determine if MAP3K1 loss-of-function enhanced PI3K inhibitor sensitivity, which could not be demonstrated. However, we instead found that breast cancers with
MAP3K1/PIK3CA co-mutations exhibit a strong luminal A phenotype. Patient-derived xenografts (PDXs) from a variety of solid tumor types showed enhanced responses to both buparlisib and
alpelisib when they harbored luminal A-like gene expression. Finally, in both buparlisib and alpelisib-treated ER+ breast cancer patients, luminal A status was associated with a superior
CBR. These data suggest that luminal A status and or co-occurring PIK3CA/MAP3K1 mutants may be predictive biomarkers for PI3K inhibition in ER+ breast cancer. RESULTS GENOMIC CORRELATES OF
OUTCOME FOLLOWING BUPARLISIB AND LETROZOLE To determine potential genomic correlates with clinical benefit to buparlisib and letrozole in metastatic ER+ breast cancer patients, we performed
MSK-IMPACT targeted next generation sequencing (tNGS) profiling for 341 genes using pre-study biopsy (where available) material or archival tissue from 33 patients subsequently enrolled and
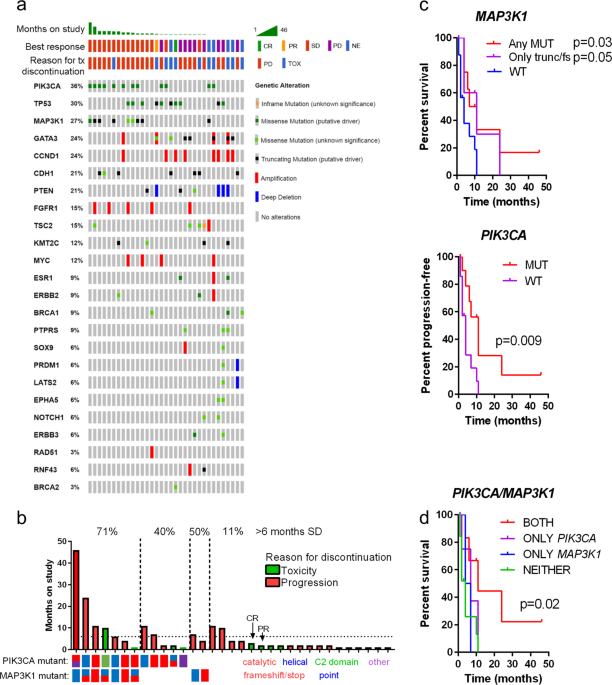
treated according to the study protocol. Mutations, amplifications, and deep deletions identified by tNGS, organized by time-on-study are shown in Fig. 1a. No genomic lesions were associated
with an objective response (partial response PR/CR), as there were only one of each in this cohort. _PTEN_ deletions or mutations were associated with PD at first scan, but a deep deletion
at the _PTEN_ locus was also detected in the only patient to achieve a PR. Interestingly, alterations in _PIK3CA were moderately_ enriched in patients with CR/PR/SD versus PD (_p_ = 0.09,
fisher’s exact) while MAP3K1 alterations were significantly enriched in patients with CR/PR/SD (_p_ = 0.01, fisher’s exact). Mutations in _PIK3CA_ were primarily hotspot mutations, while
_MAP3K1_ alterations were primarily truncating or frameshift mutations. The distribution of mutations in both MAP3K1 and PIK3CA are shown in Supplementary Fig. 1. MAP3K1 is a member of the
mitogen-activated protein kinase kinase kinase (MAP3K) family of serine/threonine kinases. The downstream effector of MAP3K1 activity (via MAP2K4) is JNK. Interestingly, MAP3K1 is distinct
from most other human kinases in that it simultaneously demonstrates E3-ubiquitin ligase activity which degrades both c-Jun and ERK1/2 via ubiquitination.12 MAP3K1 has been shown to play a
role in cell migration and survival while cleavage via caspase activation promotes apoptosis. Several genomic studies in breast cancer have revealed that MAP3K1 alterations are most common
in the luminal A subtype of breast cancer.13,14,15,16 The pattern of mutations observed both in our study (Supplementary Fig. 1) and in large-scale genomic efforts16 suggest
loss-of-function, due to the predominance of truncating and frameshift alterations. Importantly, recent data suggest that MAP3K1 deletion or loss-of-function can activate PI3K through IRS1,
though in preclinical studies, this led to resistance, not enhanced sensitivity to PI3K or AKT inhibition.17 Next, we examined the association of _PIK3CA_ and _MAP3K1_ alterations with
clinical benefit. CBR tracked with alterations in both genes; 2/5 (40%) of patients with alterations in only _PIK3CA_ and 1/2 (50%) patients with only _MAP3K1_ mutations derived clinical
benefit, whereas 5/7 (71%) patients with tumors harboring both alterations did so. Among these, two patients had SD lasting for 2 years. In contrast, patients with cancers lacking a mutation
in either gene had a CBR of only 11%, but both the single CR and PR belonged to this group (Fig. 1b). Both of these patients, despite achieving an objective response, had to come off study
prior to 6 months due to toxicity. Since it cannot be determined whether patients who were withdrawn from the study due to toxicity would have continued to benefit or progress, withdrawal
from the study for this reason was used as a censor. Using toxicity as censoring variable, stratifying progression-free survival (PFS) by presence vs. absence of a _MAP3K1_ alteration or a
_PIK3CA_ alteration both yielded improved survival (_p_ = 0.03 and _p_ = 0.009, respectively, log-rank test; Fig. 1c). Importantly, patients harboring truncating or frameshift mutations
only, as opposed to point mutations, for which biological consequences are less clear, continued to have improved outcomes (_p_ = 0.05; Fig. 1c). However, the longest-responding patient,
whose tumor harbored a MAP3K1 L380S mutation, was excluded in this analysis, despite the fact that this is a recurrent mutation in breast cancer (AACR GENIE data v4.1), suggesting it is a
loss-of-function mutation. Patients with both PIK3CA and MAP3K1 alterations appeared to achieve the greatest benefit from buparlisib and letrozole relative to patients with an alteration in
only one or the other (Fig. 1d). PI3K INHIBITION ACTIVATES JNK/CJUN SIGNALING VIA MAP3K1 We hypothesized that loss of function of MAP3K1 would render ER+ breast cancer cells more sensitive
to PI3K inhibition, potential explaining the patient outcome data. The JNK/cJUN pathway, downstream of MAP3K1, is a stress-associated pathway that has been implicated as a therapeutic escape
mechanism in cancer cells.18 Thus, we first tested the effect of buparlisib on _PIK3CA_mut ER+ T47D breast cancer cells. T47D cells were selected because they express high _MAP3K1_ mRNA
levels and an activating _PIK3CA_ mutation. Of note, we were unable to identify any ER+ breast cancer cell line within the Cancer Cell Line Encyclopedia19 with both genomic loss/inactivating
mutations in _MAP3K1_ coincident with a _PIK3CA_ mutation (Supplementary Fig. 2). Treatment with buparlisib demonstrated sustained suppression of S473 p-AKT over a 5-day period in culture.
However, as hypothesized, JNK activation measured by S73 p-cJUN was strongly activated in reciprocal fashion (Fig. 2a, b). To determine whether this upregulation of p-cJun was dependent on
MAP3K1, we generated T47D cells stably expressing short-hairpin RNA (shRNA) targeting _MAP3K1_ (shMAP3K1) or a non-targeting control (shCONTROL). Due to difficulties in identifying
effective, reproducible, and robust antibodies to reliably detect MAP3K1 protein, we used qRT-PCR to evaluate knockdown. Approximately 70% mRNA knockdown was achieved with both shRNA
sequences used, with the second sequence (shMAP3K1-03) resulting in slightly better suppression (Fig. 2c). MAP3K1 RNA interference demonstrated suppression of p-cJUN induction after 5 days
with buparlisib, without affecting baseline p-AKT levels or altering buparlisib-mediated suppression of p-AKT (Fig. 2d–f). MAP3K1 LOSS DOES NOT ALTER SENSITIVITY TO PI3K INHIBITION Since
inhibition of PI3K resulted in cJUN activation, and activation of cJUN was reduced by MAP3K1 loss, we reasoned that loss of MAP3K1 would enhance sensitivity to PI3K inhibitors by eliminating
a potential mechanism of adaptation and/or drug resistance. To test this, shMAP3K1 T47D cells were treated across a dose range of buparlisib to determine sensitivity. However, MAP3K1
knockdown did not alter the therapeutic window of buparlisib in T47D in full serum media, or estrogen-deprived conditions (Figs. 2g and 3h, respectively). Additional low density clonogenic
assays and cell proliferation in monolayer confirmed this lack of an effect of MAP3K1 loss (Supplementary Fig. 3a). A moderate decrease in shMAP3K1 cell number was observed with buparlisib
treatment, but these cells also grew more slowly in the absence of buparlisib (Supplementary Fig. 3b); after controlling for this effect there was no change in buparlisib sensitivity
(Supplementary Fig. 3c). Furthermore, transduction of _MAP3K1_ siRNA into MCF7, BT483, and T47D cells failed to impact sensitivity to buparlisib in full serum-containing media and in
estrogen-deprived conditions (Supplementary Fig. 4). Thus, despite a molecular effect of MAP3K1 loss on cJUN activation following PI3K inhibition, there was insufficient evidence to suggest
that cJUN activation promoted escape from PI3K antagonists. An alternative hypothesis could be that MAP3K1 loss increases activation of the PI3K pathway, resulting in increased dependence on
this pathway. However, we did not observe changes in PI3K activation status (p-AKT) with MAP3K1 loss, or during treatment with buparlisib (Figs. 2d and f). Furthermore, in the TCGA breast
cancer dataset, tumors with simultaneous loss of MAP3K1 or MAP2K4, its putative downstream effector, and PIK3CA alterations did not demonstrate enhanced transcriptional readouts (PIK3CA-GS,
see ref. 20) of PI3K pathway activation (Supplementary Fig. 5). LUMINAL A GENE EXPRESSION ASSOCIATES WITH RESPONSE TO BUPARLISIB AND LETROZOLE Large genomic studies have shown that luminal A
tumors are more likely to harbor both _PIK3CA_ and _MAP3K1_ alterations. Thus, we reasoned that buparlisib may elicit preferential clinical activity against this molecular subtype of breast
cancer. We speculated that this could conceivably arise from a differential dependency of more well-differentiated (i.e. luminal A) tumor cells on PI3K signaling compared to tumors that are
more de-differentiated (luminal B and others). We first tested this hypothesis using PAM50 analysis of the TCGA breast cancer data (_n_ = 959). Using correlation of the expression of each
tumor to the luminal A and luminal B PAM50 centroids, we found a strong correlation of tumors with both _MAP3K1_ (mutations/deletions) and _PIK3CA_ mutations with a luminal A-like expression
pattern vs. a luminal B expression (_p_ < 0.0001; Fig. 3a). This was unique to the tumors harboring both mutations, and was not identified in PIK3CA-only mutated or _MAP3K1_-only altered
tumors. Thus, co-occurrence of _MAP3K1_ and _PIK3CA_ mutations is associated with a strong luminal A-like phenotype in ER+ breast cancer. Based on these findings, we isolated RNA from a
subset (_n_ = 14) of available tumors from patients in the buparlisib trial as well as in 12 tumors from patients in a subsequent study of alpelisib plus letrozole.21 Using a custom-designed
NanoString codeset targeting the PAM50 geneset, we ascertained the molecular subtype by testing the association to luminal A or luminal B PAM50 centroids. Interestingly, all three patients
with clinical benefit to buparlisib/letrozole had a luminal A-like gene expression pattern in their cancer. Of these, only one had mutations in both _MAP3K1_ and _PIK3CA_, while one had only
a _MAP3K1_ alteration and the third had no alterations detected in either gene (Fig. 3b). The alpelisib trial was heavily enriched with patients with _PIK3CA_ mutations but contained no
patients with _MAP3K1_ mutations. Five of six available tumors from patients who derived clinical benefit to alpelisib/letrozole were of luminal A subtype. Overall in the combined analysis
of both trials, there was a trend toward significance (_p_ = 0.07, 1-tailed Fisher’s exact) for an association of luminal A status with clinical benefit to PI3K inhibition plus estrogen
suppression with letrozole. Thus, these data support the hypothesis that a luminal A lineage may predict benefit of PI3K-targeted inhibitors in ER+ disease, but this association should be
confirmed in a larger clinical trial. To provide additional evidence to this end, we leveraged a large database of PDXs22 from a variety of tumor types, treated with buparlisib or alpelisib.
We asked whether PAM50 subtyping of the PDXs, which has been applied to other tumor types outside of breast cancer,23,24 or the presence of genomic alterations in _MAP3K1_ or _PIK3CA_ were
associated with an improved response in vivo to PI3K inhibition. Characteristics of the PDX models analyzed are available as Supplementary Dataset 1. The response (change in tumor volume at
day 28 after starting treatment, or last time point if <28 days) in each PDX (_n_ = 1 per model per treatment22) was stratified by _MAP3K1_ alterations (mutations or deletions), _PIK3CA_
mutations, or luminal A status versus Luminal B versus all others, as predicted by a scaled-data correlation (RNAseq FPKM) to the PAM50 centroids (Fig. 4). Interestingly, PDXs with _MAP3K1_
alterations were more sensitive to alpelisib (one-tailed _p_ = 0.001), but not buparlisib. _PIK3CA_-mutant PDX models showed a trend toward an improved response to alpelisib as well
(one-tailed _p_ = 0.07). However, luminal A status predicted response to both drugs, albeit with a greater number of PDXs harboring luminal A gene expression than having alterations in
_PIK3CA_ or _MAP3K1_. The PIK3CA-GS20 gene signature was also correlated with improved response to both buparlisib and alpelisib (_p_ = 0.03 and _p_ = 0.02, respectively; Supplementary Fig.
6). Importantly, no endocrine therapy was used in this study, which contrasts with the treatment of patients on the clinical trials. However, in both clinical trials21,25 the treatment
population was endocrine-refractory tumors, and the argument could be made that the benefit of endocrine therapy on these tumors is unclear. Nonetheless, these data support the notion that
luminal A status may be a biomarker for response to PI3K inhibitors, that may also underlie the association of _MAP3K1_ alterations with clinical response to buparlisib. DISCUSSION In this
study, we explored molecular correlates of clinical benefit in a phase Ib trial of buparlisib and letrozole. We found that genomic markers of luminal A breast cancer were enriched in tumors
from patients who derived clinical benefit from the combination. Specifically, those patients with both _MAP3K1_ and _PIK3CA_ alterations appeared to derive the greatest benefit, over those
patients with one mutation or none in either gene. It is important to note that a similar association between _MAP3K1_ and _PIK3CA_ alterations with outcome on alpelisib were independently
identified in a preliminary report from ASCO 2018.26 Although there are clear caveats to modeling the molecular effects of loss-of-function of _MAP3K1_ with _PIK3CA_ mutations in breast
cancer cell lines, we did not observe a direct effect of _MAP3K1_ loss on PI3K inhibitor sensitivity in vitro. Thus, it is unclear whether MAP3K1 alterations are causal to enhanced
sensitivity to PIK3CA antagonists, or if they instead mark a specific tumor phenotype. Consistent with this second hypothesis, we found that ER+ tumors harboring both MAP3K1 and PIK3CA
alterations demonstrate a strong propensity toward a luminal A phenotype. Luminal A like tumors express higher ER levels, are more likely to co-express progesterone receptor (PR) and usually
have a low basal proliferative rate. In addition, luminal A like tumors are more sensitive to endocrine therapy, have improved prognosis, and may maintain a more differentiated state. In
patient tumors where tissue was available, we compared the gene expression patterns to luminal A/B centroids and found that patients with tumors with a luminal A expression pattern were more
likely to derive clinical benefit than patients with luminal B cancers. Furthermore, clinical benefit was observed in patients with luminal A tumors that lacked _PIK3CA_ (or _MAP3K1_)
mutations. However, it is important to note that these observations were limited to a very small sample size and should be considered anecdotal only. Several other trials have examined
molecular correlates with response to endocrine therapy and PI3K inhibition. Although the FERGI trial, which compared the pan-PI3K inhibitor pictilisib plus fulvestrant to placebo plus
fulvestrant in patients with metastatic ER+ was largely negative, patients with PR+ tumors benefitted from pictilisib/fulvestrant.27 Since PR+ patients are commonly of the luminal A
molecular subtype, these data are consistent with our own. However, in a pre-operative window trial of pictilisib and anastrozole vs. anastrozole alone, treatment with pictilisib
preferentially reduced tumor Ki67 levels, a marker of proliferation, in luminal B rather than luminal A cancers.28 While these results would appear to oppose those presented in our study,
there are likely confounding effects in this analysis; luminal A tumors typically have a very low basal proliferation rate, and are therefore much less likely to demonstrate significant
reduction in Ki67 following therapy compared to luminal B tumors. However, it is nonetheless interesting that two separate studies have found luminal A/B phenotypes involved in the response
to PI3K inhibitors in ER+ breast cancer. There are a number of caveats to the current study. First, in large comprehensive genomic studies, such as the TCGA,16 MAP3K1 mutations were not
associated with loss or reduced mRNA expression. This suggests that one of the following is true: (a) the mutations do not result in nonsense-mediate mRNA decay, but are not translated; (b)
the mutations do not result in nonsense-mediate mRNA decay, are translated, but produce non-functional proteins; or (c) there is neomorph function to MAP3K1 mutations in breast cancer that
our studies do not recapitulate, which would require more in-depth knock-in mutation studies. Next, we were unable to identify existing cell line models bearing prototypic MAP3K1 mutations.
We believe this to be a function of the frequent luminal A status of these tumors and is consistent with the general observation that Luminal A tumors are incompatible with cell line
culture, or in many cases, PDX model generation. There is another possible explanation to the association of _MAP3K1/PIK3CA_ alterations with benefit from PI3K inhibition. Luminal A tumors
typically have reduced proliferation rates and improved outcomes. Since _MAP3K1_ and _PIK3CA_ mutations were associated with prolonged stable disease, rather than objective response, it is
possible that these tumors were simply growing indolently and, thus appeared to benefit from treatment to a greater degree. A placebo control arm in a randomized trial would be required to
address such a concern. Nonetheless, our findings suggest a possible connection of luminal A tumors with response to endocrine therapy combined with a PI3K inhibitor, and support further
examination of the breast cancer molecular subtype, as well as MAP3K1 alterations, as a biomarker predictive of response to PI3K inhibitors in large randomized trials with this class of
drugs. METHODS PATIENTS The patients described in this study had histologically confirmed ER-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer refractory to
at least one line of endocrine therapy in the metastatic setting or diagnosed with metastatic breast cancer during or within 1 year of adjuvant endocrine therapy. All patients were treated
with buparlisib or alpelisib as published previously.21,25 Approval was obtained from the ethics committees (IRB# 101057, Vanderbilt University Medical Center) at the participating
institutions and regulatory authorities. All patients gave written informed consent. The study followed the Declaration of Helsinki and Good Clinical Practice guidelines. NEXT GENERATION
SEQUENCING Next generation sequencing for 341 genes was performed using the MSK-IMPACT platform on DNA isolated from formalin-fixed, paraffin-embedded surgical, or metastatic biopsies, where
available. All samples were procured prior to PI3K inhibitor treatment. The MSK-IMPACT platform and sequencing methods are described elsewhere.29 The characteristics of samples used for NGS
are presented in Supplementary Table 1. CELL CULTURE AND INHIBITORS All cell lines were purchased from ATCC. MCF7 and T47D cells were maintained in DMEM (GIBCO) supplemented with 10% fetal
bovine serum (FBS; GIBCO). BT483 cells were maintained in ATCC-formulated RPMI (ATCC, Catalog no. 30-2001) supplemented with 0.01 mg/ml bovine insulin and 20% FBS. For estrogen withdrawal
studies, cells were washed twice in serum-free phenol red-free media prior to addition of fresh phenol red-free DMEM supplemented with 10% charcoal-stripped FBS. NVP-BKM120 was purchased
from Selleckchem and solubilized in DMSO at a stock concentration of 10 mM. siRNAs against MAP3K1 were purchased from Ambion and SMARTvector shRNA were purchased from Dharmacon. Lentiviral
production was performed in 293FT cells. Cells were transduced and antibiotic selected as previously described.30 IMMUNOBLOT ANALYSIS Immunoblot analysis was carried out as described.31
Primary antibodies used were: Calnexin (#2433), p-cJUN serine 73 (#3270), cJUN (#9165), ERK (#9102), pAKT serine 473 (#4060), p-S6 (#4858) (all from Cell Signaling. 1:1000), and MAP3K1
(ab220416, Abcam; 1:100). Band intensities were quantitated using ImageJ. All pictured blots derive from the same experiment and were processed in parallel. REAL-TIME PCR Cells were
harvested and total RNA was isolated using the Maxwell nucleic isolation machine (ProMega). For real-time PCR analysis, cDNA was synthesized from total RNA using the SensiFast cDNA Synthesis
Kit (Bioline). The resulting cDNA was subjected to PCR analysis with gene-specific primers using SsoAdvanced Universal SYBR Green Supermix (BioRad). The housekeeping gene β-Actin was used
as the internal control and normalized according to the ddCT method. Primers for MAP3K1 are forward: 5′-tgatgtatggagtgttggctg-3′ and reverse 5′-aatgtgaagggatcgatggag-3′ and β-Actin are
forward 5′-agaaaatctggcaccacacc-3′ and reverse 5′-ggggtgttgaaggtctcaaa-3′. CELL VIABILITY ASSAYS MCF7, T47D, or BT483 cells were plated at a density of 10,000 cells per well in a 96-well
plate and treated with a two-fold dilution series of BKM120 for 5 days. Viability was ascertained with sulfarhodamine B (SRB) (ACROS). In brief, cells were fixed with 10% trichloroacetic
acid (TCA) at 4 °C for 30 min then stained with 0.4% SRB at room temperature for 10 min. Plates were air-dried, then SRB re-solubilized with 10 mM Tris–HCl, pH 7.5 and quantified by
absorbance (490 nm) and normalized to control (DMSO treated). For long-term assays (11 days), cells were carried out in 12-well dishes at a seeding density of 10,000 cells/well. Growth
Assays were quantified using a Coulter Counter cell counter. For clonogenic growth assays cells were plated at 1000 cells in a six-well plate. Media and drug were changed every 3 days for 30
days and harvested by fixing with 0.5% crystal violet for 15 min. Monolayer staining intensities were then quantified using a LICOR Odyssey infrared plate reader. NCOUNTER PAM50 ANALYSIS
NanoString analysis was performed on human tumors using the Elements kit. Briefly, single cross sections of FFPE tumors harvested from patient samples were used for RNA preparation using the
Maxwell-16 FFPE RNA kit (ProMega, Madison WI). Fifty nanograms of RNA (>300 nt) were used for input into the nanoString Custom Elements to detect 70 targets corresponding to 50 mRNAs of
interest and 20 housekeeping genes. Quality-control measures and normalization of data were performed using the nSolver analysis package and Log-2 transformed data were used to test
correlations with PAM50 centroids32 using the genefu package in R.33 The characteristics of samples used for PAM50 subtyping are presented in Supplementary Table 1. Normalized linear count
data for the 50 genes are available in Supplementary Dataset 2. PUBLICLY AVAILABLE DATASETS AND GENE SET ANALYSIS CCLE ANALYSIS Data for cell lines was obtained from the Cancer Cell Line
Encyclopedia assessed through www.cbioportal.org.19 TCGA ANALYSIS RNA sequencing FPKM level 3 data were downloaded from the TCGA breast cancer data portal.16 Log-2-transformed data were used
to test correlations with PAM50 centroids32 using the genefu package in R.33 Data for mutational analysis of MAP3K1 or PIK3CA in both cell lines were downloaded from the cBio portal.34
PIK3CA-GS ANALYSIS TCGA mutational data and RNA sequencing data was used to determine the PIK3CA-GS score as previously described.20 Briefly, the _z_ score of the PIK3CA-GS score (the sum of
the expression levels of the genes up-regulated in the mutant tumors minus the sum of the expression levels of the genes up-regulated in WT tumors (_n_ = 277 genes)) was used to compare
between mutational groups. PATIENT-DERIVED XENOGRAFT INHIBITOR DATA PDXs and drug treatment data were accessed from the Supplementary data file of the report.22 PDXs with mutational status
and drug sensitivity were compared across genotypes (_MAP3K1_ mutations/deletions or _PIK3CA_ mutations, excluding truncation mutations). The change in tumor volume at day 28 for each model
treated with buparlisib or alpelisib was used, or the last time point if earlier than day 28. RNA FPKM data were used to predict PAM50 subtype, using the pam50.scale model in the genefu R
package. STATISTICS Statistical analysis was performed in R (https://CRAN.R-project.org) and Graphpad Prism. REPORTING SUMMARY Further information on research design is available in the
Nature Research Reporting Summary. DATA AVAILABILITY The data generated and analyzed during this study are described in the following data record:
https://doi.org/10.6084/m9.figshare.9700373.35 Datasets in Prism file format (and accompanying.txt file versions of these datasets) supporting Figs. 1–4 of this published article, are
publicly available in the figshare repository at https://doi.org/10.6084/m9.figshare.9700373.35 Next-generation sequencing MSK-IMPACT data generated during this study, are publicly available
in cBioPortal at https://identifiers.org/cbioportal:brca_mskcc_2019. NanoString normalized linear count data generated in this study are publicly available in Supplementary dataset 2 of the
published article. The datasets analyzed during this study are publicly available in cBioPortal at https://identifiers.org/cbioportal:cellline_ccle_broad (CCLE analysis) and at
https://identifiers.org/cbioportal:brca_tcga_pub (TCGA analysis and PIK3CA-GS analysis). Patient-derived xenograft and drug treatment data analyzed in this study are publicly available in
supplementary dataset 1 of the published article. REFERENCES * Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. _CA: Cancer J. Clin._ 69, 7–34 (2019). Google Scholar *
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. _Nature_ 490, 61–70 (2012). * Bachman, K. E. et al. The PIK3CA gene is mutated with high frequency in
human breast cancers. _Cancer Biol. Ther._ 3, 772–775 (2004). Article CAS Google Scholar * Saal, L. H. et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and
ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. _Cancer Res._ 65, 2554–2559 (2005). Article CAS Google Scholar * Stemke-Hale, K. et al. An integrative genomic
and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. _Cancer Res._ 68, 6084–6091 (2008). Article CAS Google Scholar * Liu, P. et al. Oncogenic PIK3CA-driven mammary
tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. _Nat. Med._ 17, 1116–1120 (2011). Article CAS Google Scholar * Adams, J. R. et al. Cooperation
between Pik3ca and p53 mutations in mouse mammary tumor formation. _Cancer Res._ 71, 2706–2717 (2011). Article CAS Google Scholar * Fruman, D. A. et al. The PI3K pathway in human
disease. _Cell_ 170, 605–635 (2017). Article CAS Google Scholar * Hopkins, B. D. et al. Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. _Nature_ 560, 499–503
(2018). Article CAS Google Scholar * Baselga, J. et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced
breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. _Lancet Oncol._ 18, 904–916 (2017). Article CAS Google Scholar * Juric, D. et al. Phase I
dose-escalation study of taselisib, an oral PI3K inhibitor, in patients with advanced solid tumors. _Cancer Discov._ 7, 704–715 (2017). Article CAS Google Scholar * Pham, T. T., Angus, S.
P. & Johnson, G. L. MAP3K1: genomic alterations in cancer and function in promoting cell survival or apoptosis. _Genes Cancer_ 4, 419–426 (2013). Article Google Scholar * Banerji, S.
et al. Sequence analysis of mutations and translocations across breast cancer subtypes. _Nature_ 486, 405–409 (2012). Article CAS Google Scholar * Ellis, M. J. et al. Whole-genome
analysis informs breast cancer response to aromatase inhibition. _Nature_ 486, 353–360 (2012). Article CAS Google Scholar * Stephens, P. J. et al. The landscape of cancer genes and
mutational processes in breast cancer. _Nature_ 486, 400–404 (2012). Article CAS Google Scholar * TCGA. Comprehensive molecular portraits of human breast tumours. _Nature_ 490, 61–70
(2012). Article Google Scholar * Avivar-Valderas, A. et al. Functional significance of co-occurring mutations in PIK3CA and MAP3K1 in breast cancer. _Oncotarget_ 9, 21444–21458 (2018).
Article Google Scholar * Xue, Z. et al. MAP3K1 and MAP2K4 mutations are associated with sensitivity to MEK inhibitors in multiple cancer models. _Cell Res._ 28, 719–729 (2018). Article
CAS Google Scholar * Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. _Nature_ 483, 603–607 (2012). Article CAS Google
Scholar * Loi, S. et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. _Proc. Natl Acad. Sci. USA_
107, 10208–10213 (2010). Article CAS Google Scholar * Mayer, I. A. et al. A Phase Ib study of alpelisib (BYL719), a PI3Kalpha-specific inhibitor, with letrozole in ER+/HER2− metastatic
breast cancer. _Clin. Cancer Res._ 23, 26–34 (2017). Article CAS Google Scholar * Gao, H. et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial
drug response. _Nat. Med._ 21, 1318–1325 (2015). Article CAS Google Scholar * Siegfried, J. M. et al. Expression of PAM50 genes in lung cancer: evidence that interactions between hormone
receptors and HER2/HER3 contribute to poor outcome. _Neoplasia_ 17, 817–825 (2015). Article CAS Google Scholar * Zhao, S. G. et al. Associations of luminal and basal subtyping of
prostate cancer with prognosis and response to androgen deprivation therapy. _JAMA Oncol._ 3, 1663–1672 (2017). Article Google Scholar * Mayer, I. A. et al. Stand up to cancer phase Ib
study of pan-phosphoinositide-3-kinase inhibitor buparlisib with letrozole in estrogen receptor-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer. _J. Clin.
Oncol._ 32, 1202–1209 (2014). Article CAS Google Scholar * Dawson, S.-J. et al. Plasma and tumor genomic correlates of response to BYL719 in PI3KCA mutated metastatic ER-positive breast
cancer (ER+/HER2- BC). _J. Clin. Oncol._ 36, 1055–1055 (2018). Article Google Scholar * Krop, I. E. et al. Pictilisib for oestrogen receptor-positive, aromatase inhibitor-resistant,
advanced or metastatic breast cancer (FERGI): a randomised, double-blind, placebo-controlled, phase 2 trial. _Lancet Oncol._ 17, 811–821 (2016). Article CAS Google Scholar * Schmid, P. et
al. Phase II randomized preoperative window-of-opportunity study of the PI3K inhibitor pictilisib plus anastrozole compared with anastrozole alone in patients with estrogen
receptor-positive breast cancer. _J. Clin. Oncol._ 34, 1987–1994 (2016). Article CAS Google Scholar * Cheng, D. T. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of
Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. _J. Mol. Diagn._ 17, 251–264 (2015).
Article CAS Google Scholar * Balko, J. M. et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable
therapeutic targets. _Cancer Discov._ 4, 232–245 (2014). Article CAS Google Scholar * Balko, J. M., Jones, B. R., Coakley, V. L. & Black, E. P. MEK and EGFR inhibition demonstrate
synergistic activity in EGFR-dependent NSCLC. _Cancer Biol. Ther._ 8, 522–530 (2009). Article CAS Google Scholar * Parker, J. S. et al. Supervised risk predictor of breast cancer based on
intrinsic subtypes. _J. Clin. Oncol._ 27, 1160–1167 (2009). Article Google Scholar * Haibe-Kains, B. _genefu R package: Relevant Functions for Gene Expression Analysis, Especially in
Breast Cancer_. http://cran.r-project.org/web/packages/genefu/ (2009). * Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics
data. _Cancer Discov._ 2, 401–404 (2012). Article Google Scholar * Nixon, M.J. et al. _Metadata and Data Files Supporting the Related Article: PIK3CA and MAP3K1 Alterations Imply Luminal A
Status and are Associated with Clinical Benefit from pan-PI3K Inhibitor Buparlisib and Letrozole in ER+ Metastatic Breast Cancer_. https://doi.org/10.6084/m6089.figshare.9700373 (2019).
Download references ACKNOWLEDGEMENTS This work was funded by Department of Defense Era of Hope Scholar Award BC170037 (J.M.B.), R00CA181491 (J.M.B.), Susan G. Komen for the Cure Foundation
CCR14299052 (J.M.B.), AACR/Stand Up to Cancer Dream Team Translational Research Grant DT0209, Breast Cancer Specialized Program of Research Excellence (SPORE) P50 CA098131, Vanderbilt-Ingram
Cancer Center Support Grant P30 CA68485, a Breast Cancer Research Foundation grant (C.L.A.), Susan G. Komen for the Cure Foundation SAC grant (SAC100013), K23 CA127469-01A2 (I.A.M.), and by
R01 GM041890 (L.C.C.). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Medicine, Vanderbilt-Ingram Cancer Center, Vanderbilt University Medical Center, Nashville, TN, USA
Mellissa J. Nixon, Luigi Formisano, Ingrid A. Mayer & Justin M. Balko * Breast Cancer Research Program, Vanderbilt-Ingram Cancer Center, Vanderbilt University Medical Center, Nashville,
TN, USA Ingrid A. Mayer, M. Valeria Estrada, Paula I. González-Ericsson, Melinda E. Sanders & Justin M. Balko * Departments of Pathology, Microbiology, and Immunology, Vanderbilt-Ingram
Cancer Center, Vanderbilt University Medical Center, Nashville, TN, USA M. Valeria Estrada, Paula I. González-Ericsson, Melinda E. Sanders & Justin M. Balko * Department of Medicine,
Massachusetts General Hospital, Boston, MA, USA Steven J. Isakoff * University of Alabama, Birmingham, USA Andrés Forero-Torres * Memorial Sloan Kettering Cancer Center New York, New York,
NY, USA Helen Won, David B. Solit & Michael F. Berger * Weill Cornell Medical College, New York, NY, USA Lewis C. Cantley * Dana-Farber Cancer Institute, Boston, MA, USA Eric P. Winer *
Harold C. Simmons Cancer Center, UT Southwestern Medical Center, Dallas, TX, USA Carlos L. Arteaga Authors * Mellissa J. Nixon View author publications You can also search for this author
inPubMed Google Scholar * Luigi Formisano View author publications You can also search for this author inPubMed Google Scholar * Ingrid A. Mayer View author publications You can also search
for this author inPubMed Google Scholar * M. Valeria Estrada View author publications You can also search for this author inPubMed Google Scholar * Paula I. González-Ericsson View author
publications You can also search for this author inPubMed Google Scholar * Steven J. Isakoff View author publications You can also search for this author inPubMed Google Scholar * Andrés
Forero-Torres View author publications You can also search for this author inPubMed Google Scholar * Helen Won View author publications You can also search for this author inPubMed Google
Scholar * Melinda E. Sanders View author publications You can also search for this author inPubMed Google Scholar * David B. Solit View author publications You can also search for this
author inPubMed Google Scholar * Michael F. Berger View author publications You can also search for this author inPubMed Google Scholar * Lewis C. Cantley View author publications You can
also search for this author inPubMed Google Scholar * Eric P. Winer View author publications You can also search for this author inPubMed Google Scholar * Carlos L. Arteaga View author
publications You can also search for this author inPubMed Google Scholar * Justin M. Balko View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
M.J.N., L.F., J.M.B., S.J.I., A.F.-T., C.L.A., I.A.M., L.C.C. and E.W. were involved in clinical trial design and interpretation. H.W., D.S., and M.B. performed NGS and analysis and
interpretation. M.V.E., P.I.G.-E., M.E.S. performed tissue based-histological analysis and interpretation. M.J.N. and L.F. performed molecular work and bioinformatic analysis. J.M.B.
provided scientific oversight and design of the molecular and translational studies. C.L.A., M.J.N. and J.M.B. authored the paper. All authors read, approved, and participated in the writing
of the final manuscript. CORRESPONDING AUTHORS Correspondence to Carlos L. Arteaga or Justin M. Balko. ETHICS DECLARATIONS COMPETING INTERESTS J.M.B. receives consulting fees from Novartis
(not pertaining to alpelisib or buparlisib), and research support from Genentech and Incyte Pharma. M.B. receives consulting fees from Roche and research support from Illumina. I.A.M.
receives research funding from Novartis, Pfizer, and Genentech and adboards/consulting fees from Novartis, Genentech, Lilly, Astra-Zeneca, GSK, Immunomedics, Macrogenics and Seattle
Genetics. C.L.A. receives research funding from Pfizer, Lilly, Radius, and PUMA Biotechnology, stock options from Provista, and holds advisory roles with Daichii Sankyo, TAIHO Oncology,
Novartis, Merck, and Lilly. D.S. has received consulting fees from Pfizer, Loxo Oncology, Lilly Oncology, Vivideon Therapeutics and Illumina. L.C.C. owns equity in, receives compensation
from, and serves on the scientific advisory board of Agios Pharmaceuticals. He is also a founder of and receives laboratory support from Petra Pharmaceuticals and receives compensation for
being on the scientific advisory board of Petra Pharmaceuticals. No drugs from these companies were involved in this study. S.J.I. receives research funding support (to his institution) from
Genentech, PharmaMar, AstraZeneca, and Merck and receives consulting fees from Genentech, Hengrui, Puma, Immunomedics, and Myriad Genetics. The remaining authors declare no competing
interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION DATA SET 1 DATA SET 2 SUPPLEMENTARY INFORMATION REPORTING SUMMARY CHECKLIST RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0
International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the
source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative
Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by
statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Nixon, M.J., Formisano, L., Mayer, I.A. _et al._ PIK3CA and MAP3K1 alterations
imply luminal A status and are associated with clinical benefit from pan-PI3K inhibitor buparlisib and letrozole in ER+ metastatic breast cancer. _npj Breast Cancer_ 5, 31 (2019).
https://doi.org/10.1038/s41523-019-0126-6 Download citation * Received: 28 November 2018 * Accepted: 28 August 2019 * Published: 23 September 2019 * DOI:
https://doi.org/10.1038/s41523-019-0126-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
