Snord113–114 cluster maintains haematopoietic stem cell self-renewal via orchestrating the translation machinery
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

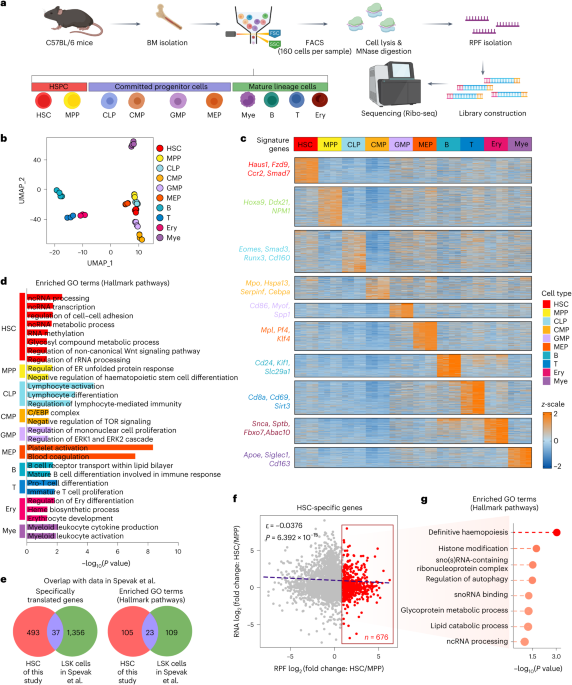
ABSTRACT Haematopoietic stem cells (HSCs) self-renew and differentiate to replenish the pool of blood cells, which require a low but finely tuned protein synthesis rate. Nonetheless, the
translatome landscape in HSCs and how the translation machinery orchestrates HSC self-renewal remain largely elusive. Here we perform ultra-low-input Ribo-seq in HSCs, progenitor and lineage
cells, and reveal HSC-specific translated genes involved in rRNA processing. We systematically profile small nucleolar RNAs (snoRNAs) and uncover an indispensable role of the SNORD113–114
cluster in regulating HSC self-renewal. Maternal knockout (Mat-KO) of this cluster substantially impairs HSC self-renewal, whereas loss of the paternal allele shows no obvious phenotype.
Mechanistically, Mat-KO results in dysregulation of translation machinery (rRNA 2′-O-Me modifications, pre-rRNA processing, 60S ribosome assembly and translation) and induces nucleolar
stress in HSCs, which exempts p53 from Mdm2-mediated proteasomal degradation and leads to apoptosis. Collectively, our study provides a promising facet to our understanding of
snoRNA-mediated regulation in HSC homeostasis. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access
through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to
this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy
now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS RNA BINDING PROTEIN SYNCRIP MAINTAINS PROTEOSTASIS AND SELF-RENEWAL OF HEMATOPOIETIC STEM AND PROGENITOR CELLS Article Open access 21 April
2023 TRNA M1A MODIFICATION REGULATE HSC MAINTENANCE AND SELF-RENEWAL VIA MTORC1 SIGNALING Article Open access 08 July 2024 YTHDC1-MEDIATED MICRORNA MATURATION IS ESSENTIAL FOR HEMATOPOIETIC
STEM CELLS MAINTENANCE Article Open access 16 October 2024 DATA AVAILABILITY All sequencing data have been deposited in the Gene Expression Omnibus under the following accession codes:
GSE166514, GSE166515, GSE166516, GSE166517, GSE166518, GSE267887, GSE269874 and GSE270936. The public mouse genome assembly GRCm38 was used for alignment. Previously published data by Spevak
et al.3 that were re-analysed here (Fig. 1e) are available under accession code GSE113886. Data supporting the findings of this study are available from the corresponding authors on
reasonable request. Source data are provided with this paper. REFERENCES * Orkin, S. H. & Zon, L. I. Hematopoiesis: an evolving paradigm for stem cell biology. _Cell_ 132, 631–644
(2008). Article CAS PubMed PubMed Central Google Scholar * Signer, R. A. J., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein
synthesis rate. _Nature_ 509, 49–54 (2014). Article CAS PubMed PubMed Central Google Scholar * Spevak, C. C. et al. Hematopoietic stem and progenitor cells exhibit stage-specific
translational programs via mTOR- and CDK1-dependent mechanisms. _Cell Stem Cell_ 26, 755–765.e7 (2020). Article CAS PubMed PubMed Central Google Scholar * Kruta, M. et al. Hsf1 promotes
hematopoietic stem cell fitness and proteostasis in response to ex vivo culture stress and aging. _Cell Stem Cell_ 28, 1950–1965.e6 (2021). Article CAS PubMed PubMed Central Google
Scholar * Lv, K. et al. HectD1 controls hematopoietic stem cell regeneration by coordinating ribosome assembly and protein synthesis. _Cell Stem Cell_ 28, 1275–1290.e9 (2021). Article CAS
PubMed PubMed Central Google Scholar * Keyvani Chahi, A. et al. PLAG1 dampens protein synthesis to promote human hematopoietic stem cell self-renewal. _Blood_ 140, 992–1008 (2022).
Article CAS PubMed PubMed Central Google Scholar * Li, C. et al. Amino acid catabolism regulates hematopoietic stem cell proteostasis via a GCN2–eIF2α axis. _Cell Stem Cell_ 29,
1119–1134.e7 (2022). Article CAS PubMed Google Scholar * Cai, X. et al. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor
cells. _Cell Stem Cell_ 17, 165–177 (2015). Article CAS PubMed PubMed Central Google Scholar * Signer, R. A. J. et al. The rate of protein synthesis in hematopoietic stem cells is
limited partly by 4E-BPs. _Genes Dev._ 30, 1698–1703 (2016). Article CAS PubMed PubMed Central Google Scholar * van Galen, P. et al. The unfolded protein response governs integrity of
the haematopoietic stem-cell pool during stress. _Nature_ 510, 268–272 (2014). Article PubMed Google Scholar * Xu, L. et al. Protein quality control through endoplasmic
reticulum-associated degradation maintains haematopoietic stem cell identity and niche interactions. _Nat. Cell Biol._ 22, 1162–1169 (2020). Article CAS PubMed PubMed Central Google
Scholar * Hidalgo San Jose, L. et al. Modest declines in proteome quality impair hematopoietic stem cell self-renewal. _Cell Rep._ 30, 69–80.e6 (2020). Article CAS PubMed Google Scholar
* Kumar, S. et al. Repolarization of HSC attenuates HSCs failure in Shwachman-Diamond syndrome. _Leukemia_ 35, 1751–1762 (2021). Article CAS PubMed Google Scholar * Warren, A. J.
Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. _Adv. Biol. Regul._ 67, 109–127 (2018). Article CAS PubMed PubMed Central Google Scholar * Woloszynek, J. R. et
al. Mutations of the SBDS gene are present in most patients with Shwachman-Diamond syndrome. _Blood_ 104, 3588–3590 (2004). Article CAS PubMed Google Scholar * Finch, A. J. et al.
Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. _Genes Dev._ 25, 917–929 (2011). Article CAS PubMed PubMed Central Google Scholar *
Wong, C. C., Traynor, D., Basse, N., Kay, R. R. & Warren, A. J. Defective ribosome assembly in Shwachman-Diamond syndrome. _Blood_ 118, 4305–4312 (2011). Article CAS PubMed Google
Scholar * Qiu, Y., Xu, M. & Huang, S. Long noncoding RNAs: emerging regulators of normal and malignant hematopoiesis. _Blood_ 138, 2327–2336 (2021). Article CAS PubMed PubMed Central
Google Scholar * Zhu, B. et al. Loss of miR-31-5p drives hematopoietic stem cell malignant transformation and restoration eliminates leukemia stem cells in mice. _Sci. Transl. Med._ 14,
eabh2548 (2022). Article CAS PubMed Google Scholar * Krivdova, G. et al. Identification of the global miR-130a targetome reveals a role for TBL1XR1 in hematopoietic stem cell
self-renewal and t(8;21) AML. _Cell Rep._ 38, 110481 (2022). Article CAS PubMed PubMed Central Google Scholar * Yang, S. et al. WDR82-binding long noncoding RNA lncEry controls mouse
erythroid differentiation and maturation. _J. Exp. Med._ 219, e20211688 (2022). Article CAS PubMed PubMed Central Google Scholar * Qian, P. et al. The Dlk1-Gtl2 locus preserves LT-HSC
function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. _Cell Stem Cell_ 18, 214–228 (2016). Article CAS PubMed Google Scholar * Watkins, N. J. & Bohnsack,
M. T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. _Wiley Interdiscip. Rev. RNA_ 3, 397–414 (2012). Article CAS
PubMed Google Scholar * McMahon, M., Contreras, A. & Ruggero, D. Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. _Wiley
Interdiscip. Rev. RNA_ 6, 173–189 (2015). Article CAS PubMed Google Scholar * Piekna-Przybylska, D., Decatur, W. A. & Fournier, M. J. The 3D rRNA modification maps database: with
interactive tools for ribosome analysis. _Nucleic Acids Res._ 36, D178–D183 (2008). Article CAS PubMed Google Scholar * Meier, U. T. The many facets of H/ACA ribonucleoproteins.
_Chromosoma_ 114, 1–14 (2005). Article CAS PubMed PubMed Central Google Scholar * Warner, W. A. et al. Expression profiling of snoRNAs in normal hematopoiesis and AML. _Blood Adv._ 2,
151–163 (2018). Article CAS PubMed PubMed Central Google Scholar * Valleron, W. et al. Specific small nucleolar RNA expression profiles in acute leukemia. _Leukemia_ 26, 2052–2060
(2012). Article CAS PubMed Google Scholar * Cohen, Y. et al. The increased expression of 14q32 small nucleolar RNA transcripts in promyelocytic leukemia cells is not dependent on
PML-RARA fusion gene. _Blood Cancer J._ 2, e92 (2012). Article CAS PubMed PubMed Central Google Scholar * Teittinen, K. J. et al. Expression of small nucleolar RNAs in leukemic cells.
_Cell. Oncol._ 36, 55–63 (2013). Article CAS Google Scholar * Ronchetti, D. et al. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. _BMC Med. Genet._ 6, 27 (2013).
Google Scholar * Sloan, K. E. et al. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. _RNA Biol._ 14, 1138–1152 (2017). Article
PubMed Google Scholar * Decatur, W. A. & Fournier, M. J. rRNA modifications and ribosome function. _Trends Biochem. Sci._ 27, 344–351 (2002). Article CAS PubMed Google Scholar *
Bellodi, C. et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. _Cancer Res._ 70, 6026–6035 (2010). Article CAS
PubMed PubMed Central Google Scholar * Bellodi, C., Kopmar, N. & Ruggero, D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis
congenita. _EMBO J._ 29, 1865–1876 (2010). Article CAS PubMed PubMed Central Google Scholar * Bellodi, C. et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding
RNA modifications in hematopoietic stem cell differentiation. _Cell Rep._ 3, 1493–1502 (2013). Article CAS PubMed Google Scholar * Guzzi, N. et al. Pseudouridylation of tRNA-derived
fragments steers translational control in stem cells. _Cell_ 173, 1204–1216.e26 (2018). Article CAS PubMed Google Scholar * Guzzi, N. et al. Pseudouridine-modified tRNA fragments repress
aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. _Nat. Cell Biol._ 24, 299–306 (2022). Article CAS PubMed PubMed Central Google Scholar *
Grozdanov, P. N., Fernandez-Fuentes, N., Fiser, A. & Meier, U. T. Pathogenic NAP57 mutations decrease ribonucleoprotein assembly in dyskeratosis congenita. _Hum. Mol. Genet_. 18,
4546–4551 (2009). Article CAS PubMed PubMed Central Google Scholar * Erales, J. et al. Evidence for rRNA 2′-O-methylation plasticity: control of intrinsic translational capabilities of
human ribosomes. _Proc. Natl Acad. Sci. USA_ 114, 12934–12939 (2017). Article CAS PubMed PubMed Central Google Scholar * Zhou, F. et al. AML1-ETO requires enhanced C/D box snoRNA/RNP
formation to induce self-renewal and leukaemia. _Nat. Cell Biol._ 19, 844–855 (2017). Article CAS PubMed Google Scholar * Lawo, S. et al. HAUS, the 8-subunit human Augmin complex,
regulates centrosome and spindle integrity. _Curr. Biol._ 19, 816–826 (2009). Article CAS PubMed Google Scholar * Corrigan, P. M., Dobbin, E., Freeburn, R. W. & Wheadon, H. Patterns
of Wnt/Fzd/LRP gene expression during embryonic hematopoiesis. _Stem Cells Dev._ 18, 759–772 (2009). Article CAS PubMed Google Scholar * Qian, P. et al. Retinoid-sensitive epigenetic
regulation of the Hoxb cluster maintains normal hematopoiesis and inhibits leukemogenesis. _Cell Stem Cell_ 22, 740–754.e7 (2018). Article CAS PubMed Google Scholar * Carpenter, K. A.,
Thurlow, K. E., Craig, S. E. L. & Grainger, S. Wnt regulation of hematopoietic stem cell development and disease. _Curr. Top. Dev. Biol._ 153, 255–279 (2023). Article CAS PubMed
PubMed Central Google Scholar * Singh, R. P. et al. HIF prolyl hydroxylase 2 (PHD2) is a critical regulator of hematopoietic stem cell maintenance during steady-state and stress. _Blood_
121, 5158–5166 (2013). Article CAS PubMed Google Scholar * Theilgaard-Mönch, K. et al. Transcription factor-driven coordination of cell cycle exit and lineage-specification in vivo
during granulocytic differentiation: in memoriam Professor Niels Borregaard. _Nat. Commun._ 13, 3595 (2022). Article PubMed PubMed Central Google Scholar * Simsek, T. et al. The distinct
metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. _Cell Stem Cell_ 7, 380–390 (2010). Article CAS PubMed PubMed Central Google Scholar * Pinho,
S. et al. VCAM1 confers innate immune tolerance on haematopoietic and leukaemic stem cells. _Nat. Cell Biol._ 24, 290–298 (2022). Article CAS PubMed PubMed Central Google Scholar * Rho,
S.-S. et al. Rap1b promotes notch-signal-mediated hematopoietic stem cell development by enhancing integrin-mediated cell adhesion. _Dev. Cell_ 49, 681–696.e6 (2019). Article CAS PubMed
Google Scholar * Zhou, F. et al. Tracing haematopoietic stem cell formation at single-cell resolution. _Nature_ 533, 487–492 (2016). Article CAS PubMed Google Scholar * Duker, A. L. et
al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. _Eur. J. Hum. Genet_ 18, 1196–1201 (2010).
Article CAS PubMed PubMed Central Google Scholar * Basbous, J. et al. Induction of ASAP (MAP9) contributes to p53 stabilization in response to DNA damage. _Cell Cycle_ 11, 2380–2390
(2012). Article CAS PubMed Google Scholar * Zhao, J. et al. Human hematopoietic stem cell vulnerability to ferroptosis. _Cell_ 186, 732–747.e16 (2023). Article CAS PubMed PubMed
Central Google Scholar * Lowe, T. M. & Eddy, S. R. A computational screen for methylation guide snoRNAs in yeast. _Science_ 283, 1168–1171 (1999). Article CAS PubMed Google Scholar
* Domostegui, A. et al. Impaired ribosome biogenesis checkpoint activation induces p53-dependent MCL-1 degradation and MYC-driven lymphoma death. _Blood_ 137, 3351–3364 (2021). Article
CAS PubMed PubMed Central Google Scholar * Pelletier, J. et al. Nucleotide depletion reveals the impaired ribosome biogenesis checkpoint as a barrier against DNA damage. _EMBO J._ 39,
e103838 (2020). Article CAS PubMed PubMed Central Google Scholar * Boulon, S., Westman, B. J., Hutten, S., Boisvert, F.-M. & Lamond, A. I. The nucleolus under stress. _Mol. Cell_
40, 216–227 (2010). Article CAS PubMed PubMed Central Google Scholar * Boisvert, F.-M., van Koningsbruggen, S., Navascués, J. & Lamond, A. I. The multifunctional nucleolus. _Nat.
Rev. Mol. Cell Biol._ 8, 574–585 (2007). Article CAS PubMed Google Scholar * Brooks, C. L. & Gu, W. p53 ubiquitination: Mdm2 and beyond. _Mol. Cell_ 21, 307–315 (2006). Article CAS
PubMed PubMed Central Google Scholar * Horn, H. F. & Vousden, K. H. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. _Oncogene_ 27, 5774–5784 (2008).
Article CAS PubMed Google Scholar * Lohrum, M. A. E., Ludwig, R. L., Kubbutat, M. H. G., Hanlon, M. & Vousden, K. H. Regulation of HDM2 activity by the ribosomal protein L11. _Cancer
Cell_ 3, 577–587 (2003). Article CAS PubMed Google Scholar * Xu, B., Gogol, M., Gaudenz, K. & Gerton, J. L. Improved transcription and translation with L-leucine stimulation of
mTORC1 in Roberts syndrome. _BMC Genomics_ 17, 25 (2016). Article PubMed PubMed Central Google Scholar * Ma, H. et al. _N_6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal
RNA methylation. _Nat. Chem. Biol._ 15, 88–94 (2019). Article CAS PubMed Google Scholar * van Tran, N. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112.
_Nucleic Acids Res._ 47, 7719–7733 (2019). Article PubMed PubMed Central Google Scholar * Sergeeva, O. et al. Modification of adenosine196 by Mettl3 methyltransferase in the 5′-external
transcribed spacer of 47S pre-rRNA affects rRNA maturation. _Cells_ 9, 1061 (2020). Article CAS PubMed PubMed Central Google Scholar * Bowie, M. B. et al. Hematopoietic stem cells
proliferate until after birth and show a reversible phase-specific engraftment defect. _J. Clin. Invest._ 116, 2808–2816 (2006). Article CAS PubMed PubMed Central Google Scholar *
Habibian, H. K. et al. The fluctuating phenotype of the lymphohematopoietic stem cell with cell cycle transit. _J. Exp. Med._ 188, 393–398 (1998). Article CAS PubMed PubMed Central
Google Scholar * Hammond, C. A., Wu, S. W., Wang, F., MacAldaz, M. E. & Eaves, C. J. Aging alters the cell cycle control and mitogenic signaling responses of human hematopoietic stem
cells. _Blood_ 141, 1990–2002 (2023). Article CAS PubMed Google Scholar * VanInsberghe, M., van den Berg, J., Andersson-Rolf, A., Clevers, H. & van Oudenaarden, A. Single-cell
Ribo-seq reveals cell cycle-dependent translational pausing. _Nature_ 597, 561–565 (2021). Article CAS PubMed Google Scholar * Tang, F. et al. mRNA-seq whole-transcriptome analysis of a
single cell. _Nat. Methods_ 6, 377–382 (2009). Article CAS PubMed Google Scholar * Marchand, V., Blanloeil-Oillo, F., Helm, M. & Motorin, Y. Illumina-based RiboMethSeq approach for
mapping of 2′-O-Me residues in RNA. _Nucleic Acids Res._ 44, e135 (2016). Article PubMed PubMed Central Google Scholar * Birkedal, U. et al. Profiling of ribose methylations in RNA by
high-throughput sequencing. _Angew. Chem. Int. Ed. Engl._ 54, 451–455 (2015). Article CAS PubMed Google Scholar * Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a
Bioconductor package for differential expression analysis of digital gene expression data. _Bioinformatics_ 26, 139–140 (2010). Article CAS PubMed Google Scholar * Bolger, A. M., Lohse,
M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. _Bioinformatics_ 30, 2114–2120 (2014). Article CAS PubMed PubMed Central Google Scholar * Langmead, B.
& Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. _Nat. Methods_ 9, 357–359 (2012). Article CAS PubMed PubMed Central Google Scholar * Lê, S., Josse, J. & Husson, F.
FactoMineR: an R Package for multivariate analysis. _J. Stat. Softw._ 25, 1–18 (2008). Article Google Scholar * Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner
with low memory requirements. _Nat. Methods_ 12, 357–360 (2015). Article CAS PubMed PubMed Central Google Scholar * Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to
work with high-throughput sequencing data. _Bioinformatics_ 31, 166–169 (2015). Article CAS PubMed Google Scholar * Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics
enrichment tools: paths toward the comprehensive functional analysis of large gene lists. _Nucleic Acids Res._ 37, 1–13 (2009). Article PubMed Google Scholar * Subramanian, A. et al. Gene
set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. _Proc. Natl Acad. Sci. USA_ 102, 15545–15550 (2005). Article CAS PubMed PubMed
Central Google Scholar * Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. _Bioinformatics_ 29, 15–21 (2013). Article CAS PubMed Google Scholar * Patro, R., Duggal, G., Love,
M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. _Nat. Methods_ 14, 417–419 (2017). Article CAS PubMed PubMed
Central Google Scholar * Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. _F1000Research_ 4, 1521
(2015). Article PubMed Google Scholar * Stuart, T. et al. Comprehensive integration of single-cell data. _Cell_ 177, 1888–1902.e21 (2019). Article CAS PubMed PubMed Central Google
Scholar * Chen, J., Bardes, E. E., Aronow, B. J. & Jegga, A. G. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. _Nucleic Acids Res._ 37, W305–W311
(2009). Article CAS PubMed PubMed Central Google Scholar * Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. _Bioinformatics_ 26,
841–842 (2010). Article CAS PubMed PubMed Central Google Scholar * Penzo, M., Carnicelli, D., Montanaro, L. & Brigotti, M. A reconstituted cell-free assay for the evaluation of the
intrinsic activity of purified human ribosomes. _Nat. Protoc._ 11, 1309–1325 (2016). Article CAS PubMed Google Scholar * Panthu, B., Décimo, D., Balvay, L. & Ohlmann, T. In vitro
translation in a hybrid cell free lysate with exogenous cellular ribosomes. _Biochem. J._ 467, 387–398 (2015). Article CAS PubMed Google Scholar * Schmidt, E. K., Clavarino, G., Ceppi,
M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. _Nat. Methods_ 6, 275–277 (2009). Article CAS PubMed Google Scholar * Ge, J. et al. Dyskerin ablation in
mouse liver inhibits rRNA processing and cell division. _Mol. Cell. Biol._ 30, 413–422 (2010). Article CAS PubMed Google Scholar * Wang, H. et al. SARS-CoV-2 N protein potentiates host
NPM1-snoRNA translation machinery to enhance viral replication. _Signal Transduct. Target Ther._ 7, 356 (2022). Article CAS PubMed PubMed Central Google Scholar Download references
ACKNOWLEDGEMENTS This work was supported by grants from the National Key Research and Development Program of China (2022YFA1103500 and 2024YFA1107102 to P.Q., 2021YFA1102400 to F.W. and
2022YFA1106300 and 2024YFA1803500 to J.X.), the National Natural Science Foundation of China (82222003 and 92268117 to P.Q., 92268205 and 82330007 to J.Y., 82470123 to F.W. and 31900815 to
H.W.), the Key R&D Program of Zhejiang (2024SSYS0024, 2024SSYS0023 and 2024SSYS0025 to P.Q.), the Zhejiang Provincial Natural Science Foundation of China (Z24H080001 to P.Q.), the
Department of Science and Technology of Zhejiang Province (2023R01012 to P.Q.), the Fundamental Research Funds for the Central Universities (226-2024-00007 to P.Q.), Chinese Academy of
Medical Sciences Innovation Fund for Medical Sciences (2021-I2M-1-040 to F.W.) and Zhejiang Province Postdoctoral Research Excellence Funding Project (ZJ2022098 to H.W.). Haihe Laboratory of
Cell Ecosystem Innovation Fund (22HHXBSS00027 to J.Y.). P.Q. gratefully acknowledges the support of the K.C. Wong Education Foundation. Technical support was provided by the core facilities
at the Zhejiang University School of Medicine (special thanks to C. Guo and X. Hong for assistance during experiments), as well as the core facilities at the Zhejiang University Medical
Center and Liangzhu Laboratory. We thank State Key Laboratory of Common Mechanism Research of Major Diseases Platform for consultation and instrument availability that supported this work,
and High-performance Computing Platform at the Center for Bioinformatics, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. AUTHOR INFORMATION Author notes * These
authors contributed equally: Hui Wang, Zhaoru Zhang, Chenxi Han, Penglei Jiang, Jiayue Xu, Yingli Han. AUTHORS AND AFFILIATIONS * Bone Marrow Transplantation Center of the First Affiliated
Hospital & Liangzhu Laboratory, Zhejiang University School of Medicine, State Key Laboratory of Experimental Hematology, Hangzhou, China Hui Wang, Zhaoru Zhang, Penglei Jiang, Yingli
Han, Deyu Huang & Pengxu Qian * Center for Stem Cell and Regenerative Medicine, Zhejiang University School of Medicine, Hangzhou, China Hui Wang, Zhaoru Zhang, Penglei Jiang, Yingli Han,
Deyu Huang & Pengxu Qian * Institute of Hematology, Zhejiang University & Zhejiang Engineering Laboratory for Stem Cell and Immunotherapy, Hangzhou, China Hui Wang, Zhaoru Zhang,
Penglei Jiang, Yingli Han, Deyu Huang & Pengxu Qian * Department of Hematology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical
College, Beijing, China Chenxi Han & Jian Li * State Key Laboratory of Common Mechanism Research for Major Disease, Department of Biochemistry and Molecular Biology, Institute of Basic
Medical Sciences, Chinese Academy of Medical Sciences, School of Basic Medicine Peking Union Medical College, Beijing, China Jiayue Xu, Jia Yu & Fang Wang * The Key Laboratory of RNA and
Hematopoietic Regulation, Chinese Academy of Medical Sciences, Beijing, China Jiayue Xu, Jia Yu & Fang Wang * State Key Laboratory of Proteomics, Academy of Military Medical Sciences,
Academy of Military Sciences, Beijing, China Jie Zhou & Bing Liu * State Key Laboratory of Experimental Hematology, Institute of Hematology, Fifth Medical Center of Chinese PLA General
Hospital, Beijing, China Jie Zhou & Bing Liu * Stowers Institute for Medical Research, Kansas City, MO, USA Michael Durnin & Shiyuan Chen * Institute of Environmental Medicine, and
Cancer Center of the First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China Yaxin Liu & Jinghao Sheng * Collaborative Innovation Center for Diagnosis and
Treatment of Infectious Diseases, Zhejiang University, Hangzhou, China Yaxin Liu & Jinghao Sheng * MOE Key Laboratory of Macromolecular Synthesis and Functionalization, Department of
Polymer Science and Engineering, Zhejiang University, Hangzhou, China Jie Cao & Jianzhao Liu * Institute of Blood Transfusion, Chinese Academy of Medical Sciences, Chengdu, China Jia Yu
& Fang Wang * State Key Laboratory of Experimental Hematology, National Clinical Research Center for Blood Diseases, Institute of Hematology and Blood Diseases Hospital, Haihe Laboratory
of Cell Ecosystem, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin, China Jia Yu Authors * Hui Wang View author publications You can also search for this
author inPubMed Google Scholar * Zhaoru Zhang View author publications You can also search for this author inPubMed Google Scholar * Chenxi Han View author publications You can also search
for this author inPubMed Google Scholar * Penglei Jiang View author publications You can also search for this author inPubMed Google Scholar * Jiayue Xu View author publications You can also
search for this author inPubMed Google Scholar * Yingli Han View author publications You can also search for this author inPubMed Google Scholar * Deyu Huang View author publications You
can also search for this author inPubMed Google Scholar * Jian Li View author publications You can also search for this author inPubMed Google Scholar * Jie Zhou View author publications You
can also search for this author inPubMed Google Scholar * Michael Durnin View author publications You can also search for this author inPubMed Google Scholar * Shiyuan Chen View author
publications You can also search for this author inPubMed Google Scholar * Yaxin Liu View author publications You can also search for this author inPubMed Google Scholar * Jinghao Sheng View
author publications You can also search for this author inPubMed Google Scholar * Jie Cao View author publications You can also search for this author inPubMed Google Scholar * Jianzhao Liu
View author publications You can also search for this author inPubMed Google Scholar * Bing Liu View author publications You can also search for this author inPubMed Google Scholar * Jia Yu
View author publications You can also search for this author inPubMed Google Scholar * Fang Wang View author publications You can also search for this author inPubMed Google Scholar *
Pengxu Qian View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS H.W. and P.Q. conceived the project and designed the experiments. H.W.
performed the experiments, analysed data and wrote the manuscript. C.H. and J. Li helped perform library construction work of Ribo-seq experiments. P.J., J.X. and S.C. analysed
second-generation sequencing data. Z.Z., Y.H. and D.H. helped perform transplantation experiments and undertook mouse breeding work. Z.Z. and J.Z. performed the SMART-Seq2 experiments. Y.L.
and J.S. helped perform the northern blot experiments. J.C. and J. Liu helped perform the UHPLC–MS experiments. M.D. helped make the SNORD113–114 cluster knockout mice. P.Q., B.L., J.Y. and
F.W. supervised the overall project and co-wrote the manuscript. All authors contributed to reading and editing the manuscript. CORRESPONDING AUTHORS Correspondence to Bing Liu, Jia Yu, Fang
Wang or Pengxu Qian. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Cell Biology_ thanks the anonymous
reviewers for their contribution to the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published
maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 GATING STRATEGIES USED IN FLOW CYTOMETRY. A, Flow cytometry plots showing gating strategies used for detecting HSPCs,
progenitors, mature cells. B, Flow cytometry plots showing gating strategies used for detecting human HSC. C, Flow cytometry plots showing gating strategies used for detecting donor
repopulation rate. EXTENDED DATA FIG. 2 TRANSLATOME PROFILING OF HAEMATOPOIETIC CELLS. A, Barplot showing representative reads distributions along the transcripts. Read position relative to
different reading frames (0, 1, 2) are colour-coded. B, Barplot showing representative ratios of reads belonging to different reading frames. C, Reads length distribution frequencies of all
cell types. D, Scatterplots showing Pearson’s correlation between two batches of data from Ribo-seq. E, Scatterplots showing Pearson’s correlation between RNA level and RPF level of genes in
each cell type. Source data EXTENDED DATA FIG. 3 HSC-SPECIFIC SNORD113–114 CLUSTER IS INDISPENSABLE FOR HSC MAINTENANCE. A, Heatmap of snoRNA expression (Z-score normalized) in progenitors
and lineage mature cells. B, C, Raw read counts of different types of ncRNAs got from sncRNA-seq. D, Sequence alignments of snoRNAs belonging to SNORD113–114 cluster between _Mus musculus_
and _Homo sapiens_. Only identical sequences of specific snoRNAs between human and mouse are presented. E, Electrophoresis showing PCR products patterns got from genotyping of WT and Mat-KO
mice. F, Absolute numbers of total BM cells of WT and Mat-KO mice were studied by flow cytometry (n = 4 for WT, n = 3 for Mat-KO biological replicates). Error bars, SEM. G-J, Frequencies and
absolute numbers of SLAM HSCs (G, H) and progenitors (I, J) from WT and Mat-KO mice detected by flow cytometry (n = 4 for WT, n = 3 for Mat = -KO biological replicates). Error bars, SEM. K,
L, Frequency (l), absolute number (m) of lineage cells collected from WT and Mat-KO mice (n = 4 for WT, n = 3 for Mat-KO biological replicates). Error bars, SEM. All _P_ values were
determined by two-tailed unpaired Student’s _t_-test. Source data EXTENDED DATA FIG. 4 SNORD113–114 MAT-KO DOES NOT AFFECT THE SPLEEN HAEMATOPOIESIS. A, B, Spleen weight (A) and cellularity
(B) were examined and compared between WT and Mat-KO mice. N = 3 for WT, n = 4 for Mat-KO biological replicates in (A). N = 4 for WT, n = 3 for Mat-KO in (B). Error bars, SEM. C-H, Frequency
and absolute number of spleen HSC (C, D), progenitors (E, F) and lineage cell (G, H) were analysed and compared between WT and Mat-KO mice (n = 4 for WT, n = 3 for Mat-KO biological
replicates). Error bars, SEM. I, Whole blood counts of WT and Mat-KO mice (n = 7 biological replicates). Error bars, SEM. All _P_ values were determined by two-tailed unpaired Student’s
_t_-test. Source data EXTENDED DATA FIG. 5 SNORD113–114 PAT-KO DOES NOT LEAD TO HSC DEFICIENCY. A, BM cellularity of WT and Pat-KO mice (n = 3 biological replicates). Error bars, SEM. B, C,
Frequency (B) and absolute number (C) of HSCs were analysed and compared between WT and Pat-KO mice (n = 3 biological replicates). Error bars, SEM. D, E, Apoptosis rate (D) and cell cycle
distribution (E) were analysed and compared between WT and Pat-KO mice (n = 3 biological replicates). Different cell cycle phases are colour-coded and presented as a percentage of whole
population (n = 6 biological replicates). Error bars, SEM. F, Schematic presentation of workflow of transplantation using sorted HSCs. G, H, Repopulation rate (G) and lineage distribution
(H) of WT and Pat-KO mice HSC (n = 3 biological replicates). Error bars, SEM; All _P_ values were determined by two-tailed unpaired Student’s _t_-test. Source data EXTENDED DATA FIG. 6
SNORNAS IN SNORD113–114 CLUSTER ARE SPECIFICALLY IMPORTANT FOR HSC. A-F, SNORD116 was not functionally important for HSCs. SNORD116 were knocked out using specific sgRNAs. After 7-day’s
culture in 96-well plates, expressions of snoRNAs were examined by RT–qPCR (A), HSPC ratios and absolute numbers were examined by flow cytometry (B-D), and colony formation ability was
examined (E, F). n = 3 biological replicates. Scale bar, 200 μm. Error bars, SEM. G, Expressions of specific snoRNAs were examined by RT–qPCR and compared between HSCs from male and female
mice. n = 3 biological replicates. Error bars, SEM. H-K, SnoRNAs in SNORD113–114 cluster were also functionally important for human HSCs. CD34+ HSCs were collected from healthy donors, and 3
snoRNAs (SNORD113–6, SNORD113-9, SNORD114-1) were knocked out using specific sgRNAs. After 7-day’s culture in 96-well plates, expressions of snoRNAs were examined by RT–qPCR (H), HSPC
ratios were examined by flow cytometry (I), and colony formation ability was examined (J, K). n = 3 biological replicates. Scale bar, 200 μm. Error bars, SEM. Cartoon illustrations were
created by Figdraw. All _P_ values were determined by two-tailed unpaired Student’s _t_-test. EXTENDED DATA FIG. 7 HIGH-THROUGHPUT SEQUENCING ANALYSIS OF HSCS OF WT/MAT-KO MICE. A, Raw read
counts of different types of ncRNAs got from sncRNA-seq of WT and Mat-KO mice. B, Western blot result showing expressions of Dlk1 in WT and Mat-KO HSCs (n = 3 biological replicates). C, PCA
analysis of sncRNA-seq of WT/Pat-KO mice LSK cells. D, Volcano-plot showing differentially expressed snoRNAs between WT/Pat-KO LSK cells. E, Raw read counts of different types of ncRNAs got
from sncRNA-seq of WT and Pat-KO mice. F, Heatmap showing expression of snoRNAs belonging to SNORD113–114 cluster in WT/Pat-KO LSK cells. Only detected snoRNAs are shown. G, PCA analysis of
RNA-seq results of WT/Pat-KO HSCs. H, Volcano-plot showing differentially expressed genes between WT/Pat-KO HSCs. I, Visualization of colour-coded clustering of HSCs via t-SNE based on
experimental groups and gene signature, showing distribution of HSCs of WT and Mat-KO mice. J, Distribution of WT/Mat-KO HSCs among different cell populations. K, Violin-plot of different
marker genes, showing expression patterns in all clusters. Violin box is colour-coded as for population clusters shown in Fig. 4m. L, GO term analysis of signature genes of different
populations. Source data EXTENDED DATA FIG. 8 LOSS OF SNORD113–114 CLUSTER DYSREGULATES RRNA MODIFICATION IN HSCS. A, Read length distributions of samples from RiboMethSeq. B, PCA analysis
of RNA fragments got from RiboMethSeq of WT/Mat-KO LSK cells. C, Conservation of all differentially methylated sites detected by RiboMethSeq across _E. coli_, _S. cerevisiae_, _M. musculus_
and _H. sapiens_. Different species are colour-coded, and sites that are not conserved were made vacant in the coloured circle representing according species. D, 2′-O-Me modification levels
were examined by RTL-P. Upper panel shows schematic of primers used for reverse transcription and qPCR. Lower panel shows agarose electrophoresis images of semi-quantitative PCR results (n =
3 biological replicates). E, Targets of snoRNAs in SNORD113–114 cluster were predicted using online tool Snoscan. Predicted targets are listed under each snoRNA, and the nucleotide
predicted to be 2′-O-methylated are labelled red. F, Relative methylation ratio (from data of RiboMethSeq) of specific sites that were predicted to be targeted by snoRNAs in SNORD113–114
cluster (n = 3 biological replicates). Error bars, SEM. All _P_ values were determined by two-tailed unpaired Student’s _t_-test. Source data EXTENDED DATA FIG. 9 LOSS OF SNORD113–114
CLUSTER IMPAIRS RRNA PROCESSING AND TRANSLATION IN HSCS. A, Schematic representation of primer designing for detecting various pre-rRNA isoforms generated during pre-rRNA processing. Red
bars show PCR products of according primers. B–E, Relative expression levels of various pre-rRNA isoforms detected by RT–qPCR (n = 3 biological replicates). Error bars, SEM. F, PCA analysis
of RPFs of WT and Mat-KO HSCs from Ribo-seq. G, Reads length distribution frequencies of RPFs from Ribo-seq. H-J, Expressions of representative genes in different sucrose gradients from
polysome profiling assay were examined by RT–qPCR (n = 3 biological replicates). Error bars, SEM. K, Expressions of snoRNAs were examined by RT–qPCR after WT/Mat-KO HSCs were infected by
according viruses (n = 3 biological replicates). Error bars, SEM. L, M, BM cells were infected by lentivirus carrying vectors expressing snoRNAs or according control empty vectors, and were
analysed using flow cytometry after 7-day culture. Frequencies (L) and absolute numbers (M) were analysed in different groups. n = 3 biological replicates. Error bars, SEM. All _P_ values
were determined by two-tailed unpaired Student’s _t_-test. Source data EXTENDED DATA FIG. 10 SNORNAS IN SNORD113–114 CLUSTER FUNCTION THROUGH GUIDING 2′-O-METHYLATION OF SPECIFIC RRNA SITES.
A-D, 2′-O-Me modifications of specific sites targeted by the ectopically expressed snoRNAs were also examined by RTL-P. E-G, Reconstitution ability analysis of HSCs from WT and Mat-KO mice
after L-leucine treatment. BM cells of WT/Mat-KO mice were treated with L-leucine at dose of 5 μM for 48 h, and then were transplanted (2 e5/sample) into lethally irradiated recipient mice
together with CD45.1 competitor cells (2 e5/sample). Repopulation rate (E), frequency of TNC (F) and absolute cell number (G) were analysed and compared between WT and Mat-KO mice at 16
weeks post-transplantation using flow cytometry (n = 7 biological replicates). Error bars, SEM. H-J, Statistical analysis of apoptosis ratio, cell cycle distributions and translation rate
(OP-Puro assay) of HSCs after transplantation. N = 7 biological replicates. Error bars, SEM. K-P, 2′-O-Me modifications of specific sites were examined by RTL-P after L-leucine treatment (5
μM for 48 h). Q, p53 expressions in WT/Mat-KO HSCs after L-leucine treatment (5 μM for 48 h). R, Expressions of HSC self-renewal related genes in WT/Mat-KO HSCs after L-leucine treatment (5
μM for 48 h). n = 3 biological replicates. Error bars, SEM. All _P_ values were determined by two-tailed unpaired Student’s _t_-test. Source data SUPPLEMENTARY INFORMATION REPORTING SUMMARY
SUPPLEMENTARY TABLES Supplementary Table 1. RPFs of genes of all haematopoietic cell types by Ribo-seq. Supplementary Table 2. CPM of sncRNAs acquired from sncRNA-seq. Supplementary Table 3.
FPKMs of all transcripts and DEGs in LSK cells of WT and Mat-KO mice by RNA-seq. Supplementary Table 4. GO analysis using biological process between Mat-KO/WT group. Supplementary Table 5.
DEGs between WT and Mat-KO by single-cell RNA-seq. Supplementary Table 6. DMSs between WT and Mat-KO by RiboMethSeq. Supplementary Table 7. RPFs of genes of Mat-KO/WT HSC by Ribo-seq.
Supplementary Table 8. Information of reagents and materials used in this study. SOURCE DATA SOURCE DATA FIGS. 2–5, 7 AND 8, AND EXTENDED DATA FIGS. 2–5 AND 7–9 Combined file for statistical
source data for all figures. SOURCE DATA FIGS. 5G,K, 7D–G AND 8K, AND EXTENDED DATA FIGS. 3E, 7B, 8D AND 10A,C,K,M,Q Combined file for unprocessed western blots and gels for all figures.
RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other
rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Wang, H., Zhang, Z., Han, C. _et al._ SNORD113–114 cluster maintains haematopoietic stem cell self-renewal via orchestrating the translation
machinery. _Nat Cell Biol_ 27, 246–261 (2025). https://doi.org/10.1038/s41556-024-01593-7 Download citation * Received: 20 July 2023 * Accepted: 11 December 2024 * Published: 31 January 2025
* Issue Date: February 2025 * DOI: https://doi.org/10.1038/s41556-024-01593-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable
link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
