Bipartite interactions, antibiotic production and biosynthetic potential of the arabidopsis leaf microbiome
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Plants are colonized by phylogenetically diverse microorganisms that affect plant growth and health. Representative genome-sequenced culture collections of bacterial isolates from
model plants, including _Arabidopsis thaliana_, have recently been established. These resources provide opportunities for systematic interaction screens combined with genome mining to
discover uncharacterized natural products. Here, we report on the biosynthetic potential of 224 strains isolated from the _A. thaliana_ phyllosphere. Genome mining identified more than 1,000
predicted natural product biosynthetic gene clusters (BGCs), hundreds of which are unknown compared to the MIBiG database of characterized BGCs. For functional validation, we used a
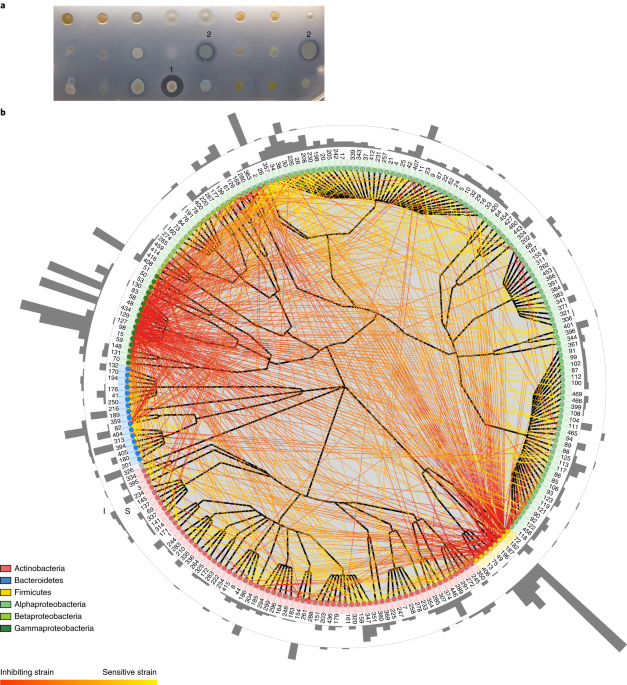
high-throughput screening approach to monitor over 50,000 binary strain combinations. We observed 725 inhibitory interactions, with 26 strains contributing to the majority of these. A
combination of imaging mass spectrometry and bioactivity-guided fractionation of the most potent inhibitor, the BGC-rich _Brevibacillus_ sp. Leaf182, revealed three distinct natural product
scaffolds that contribute to the observed antibiotic activity. Moreover, a genome mining-based strategy led to the isolation of a _trans_-acyltransferase polyketide synthase-derived
antibiotic, macrobrevin, which displays an unprecedented natural product structure. Our findings demonstrate that the phyllosphere is a valuable environment for the identification of
antibiotics and natural products with unusual scaffolds. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS
OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn
more Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant
access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions *
Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS EXPLORING THE SPECIALIZED METABOLOME OF THE PLANT PATHOGEN _STREPTOMYCES_ SP. 11-1-2 Article Open access 06
May 2024 GENOMIC AND METABOLOMIC INSIGHTS INTO THE ANTIMICROBIAL COMPOUNDS AND PLANT GROWTH-PROMOTING POTENTIAL OF _BACILLUS VELEZENSIS_ B115 Article Open access 28 March 2025 CORRELATIVE
METABOLOGENOMICS OF 110 FUNGI REVEALS METABOLITE–GENE CLUSTER PAIRS Article 06 March 2023 REFERENCES * Dewick, P. M. _Medicinal Natural Products: A Biosynthetic Approach_ 3rd edn (Wiley,
Chichester, 2009). * Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs from 1981 to 2014. _J. Nat. Prod._ 79, 629–661 (2016). Article CAS PubMed Google Scholar *
Mlot, C. Antibiotics in nature: beyond biological warfare. _Science_ 324, 1637–1639 (2009). Article CAS PubMed Google Scholar * Meiser, P., Bode, H. B. & Müller, R. The unique
DKxanthene secondary metabolite family from the myxobacterium _Myxococcus xanthus_ is required for developmental sporulation. _Proc. Natl Acad. Sci. USA_ 103, 19128–19133 (2006). Article
CAS PubMed PubMed Central Google Scholar * Hawver, L. A., Jung, S. A. & Ng, W. L. Specificity and complexity in bacterial quorum-sensing systems. _FEMS Microbiol. Rev._ 40, 738–752
(2016). Article CAS PubMed PubMed Central Google Scholar * Höfer, I. et al. Insights into the biosynthesis of hormaomycin, an exceptionally complex bacterial signaling metabolite.
_Chem. Biol._ 18, 381–391 (2011). Article CAS PubMed Google Scholar * Phelan, V. V., Liu, W. T., Pogliano, K. & Dorrestein, P. C. Microbial metabolic exchange—the
chemotype-to-phenotype link. _Nat. Chem. Biol._ 8, 26–35 (2012). Article CAS Google Scholar * Saha, R., Saha, N., Donofrio, R. S. & Bestervelt, L. L. Microbial siderophores: a mini
review. _J. Basic Microbiol._ 53, 303–317 (2013). Article PubMed Google Scholar * Wilson, M. C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire.
_Nature_ 506, 58–62 (2014). Article CAS PubMed Google Scholar * Lincke, T., Behnken, S., Ishida, K., Roth, M. & Hertweck, C. Closthioamide: an unprecedented polythioamide antibiotic
from the strictly anaerobic bacterium _Clostridium cellulolyticum_. _Angew. Chem._ 49, 2011–2013 (2010). Article CAS Google Scholar * Pidot, S. J., Coyne, S., Kloss, F. & Hertweck, C.
Antibiotics from neglected bacterial sources. _Int. J. Med. Microbiol._ 304, 14–22 (2014). Article CAS PubMed Google Scholar * Wilson, M. C. & Piel, J. Metagenomic approaches for
exploiting uncultivated bacteria as a resource for novel biosynthetic enzymology. _Chem. Biol._ 20, 636–647 (2013). Article CAS PubMed Google Scholar * Rondon, M. R. et al. Cloning the
soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. _Appl. Environ. Microbiol._ 66, 2541–2547 (2000). Article CAS PubMed PubMed
Central Google Scholar * Banik, J. J. & Brady, S. F. Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. _Curr.
Opin. Microbiol._ 13, 603–609 (2010). Article CAS PubMed PubMed Central Google Scholar * Vorholt, J. A., Vogel, C., Carlström, C. I. & Müller, D. B. Establishing causality:
opportunities of synthetic communities for plant microbiome research. _Cell Host Microbe_ 22, 142–155 (2017). Article CAS PubMed Google Scholar * Bai, Y. et al. Functional overlap of the
_Arabidopsis_ leaf and root microbiota. _Nature_ 528, 364–369 (2015). Article CAS PubMed Google Scholar * Vorholt, J. A. Microbial life in the phyllosphere. _Nat. Rev. Microbiol._ 10,
828–840 (2012). Article CAS PubMed Google Scholar * Ryffel, F. et al. Metabolic footprint of epiphytic bacteria on _Arabidopsis thaliana_ leaves. _ISME J._ 10, 632–643 (2016). Article
CAS PubMed Google Scholar * Blin, K. et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. _Nucleic Acids Res._ 45, W36–W41 (2017). Article
CAS PubMed PubMed Central Google Scholar * Medema, M. H. et al. Minimum information about a biosynthetic gene cluster. _Nat. Chem. Biol._ 11, 625–631 (2015). Article CAS PubMed PubMed
Central Google Scholar * Yang, S. C., Lin, C. H., Sung, C. T. & Fang, J. Y. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. _Front. Microbiol._ 5,
241 (2014). PubMed PubMed Central Google Scholar * Helfrich, E. J. & Piel, J. Biosynthesis of polyketides by _trans_-AT polyketide synthases. _Nat. Prod. Rep._ 33, 231–316 (2016).
Article CAS PubMed Google Scholar * Blin, K., Medema, M. H., Kottmann, R., Lee, S. Y. & Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite
biosynthetic gene clusters. _Nucleic Acids Res._ 45, D555–D559 (2017). Article CAS PubMed Google Scholar * Hillenmeyer, M. E., Vandova, G. A., Berlew, E. E. & Charkoudian, L. K.
Evolution of chemical diversity by coordinated gene swaps in type II polyketide gene clusters. _Proc. Natl Acad. Sci. USA_ 112, 13952–13957 (2015). Article CAS PubMed PubMed Central
Google Scholar * Lazos, O. et al. Biosynthesis of the putative siderophore erythrochelin requires unprecedented crosstalk between separate nonribosomal peptide gene clusters. _Chem. Biol._
17, 160–173 (2010). Article CAS PubMed Google Scholar * Lombó, F. et al. Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression
in two _Streptomyces_ species. _ChemBioChem_ 7, 366–376 (2006). Article CAS PubMed Google Scholar * Arrebola, E. et al. Mangotoxin: a novel antimetabolite toxin produced by _Pseudomonas
syringae_ inhibiting ornithine/arginine biosynthesis. _Physiol. Mol. Plant Pathol._ 63, 117–127 (2003). Article CAS Google Scholar * Bassler, B. L. & Losick, R. Bacterially speaking.
_Cell_ 125, 237–246 (2006). Article CAS PubMed Google Scholar * Pandey, S. S., Patnana, P. K., Rai, R. & Chatterjee, S. Xanthoferrin, the α-hydroxycarboxylate-type siderophore of
_Xanthomonas campestris_ pv. _campestris_, is required for optimum virulence and growth inside cabbage. _Mol. Plant Pathol._ 18, 949–962 (2017). Article CAS PubMed Google Scholar *
Barona-Gomez, F., Wong, U., Giannakopulos, A. E., Derrick, P. J. & Challis, G. L. Identification of a cluster of genes that directs desferrioxamine biosynthesis in _Streptomyces
coelicolor_ M145. _J. Am. Chem. Soc._ 126, 16282–16283 (2004). Article CAS PubMed Google Scholar * Lee, J. Y. et al. Biosynthetic analysis of the petrobactin siderophore pathway from
_Bacillus anthracis_. _J. Bacteriol._ 189, 1698–1710 (2007). Article CAS PubMed Google Scholar * Barry, S. M. & Challis, G. L. Recent advances in siderophore biosynthesis. _Curr.
Opin. Chem. Biol._ 13, 205–215 (2009). Article CAS PubMed Google Scholar * Lindow, S. E. & Brandl, M. T. Microbiology of the phyllosphere. _Appl. Environ. Microbiol._ 69, 1875–1883
(2003). Article CAS PubMed PubMed Central Google Scholar * Lindow, S. E. & Leveau, J. H. Phyllosphere microbiology. _Curr. Opin. Biotechnol._ 13, 238–243 (2002). Article CAS
PubMed Google Scholar * Schöner, T. A. et al. Aryl polyenes, a highly abundant class of bacterial natural products, are functionally related to antioxidative carotenoids. _ChemBioChem_ 17,
247–253 (2016). Article CAS PubMed Google Scholar * Wang, M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking.
_Nat. Biotechnol._ 34, 828–837 (2016). Article CAS PubMed PubMed Central Google Scholar * Mootz, H. D. & Marahiel, M. A. The tyrocidine biosynthesis operon of Bacillus brevis:
complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. _J. Bacteriol._ 179, 6843–6850 (1997). Article CAS PubMed PubMed Central Google
Scholar * Gebhardt, K., Pukall, R. & Fiedler, H. P. Streptocidins A-D, novel cyclic decapeptide antibiotics produced by _Streptomyces_ sp. Tu 6071. I. Taxonomy, fermentation, isolation
and biological activities. _J. Antibiot._ 54, 428–433 (2001). Article CAS Google Scholar * Zgurskaya, H. I., Löpez, C. A. & Gnanakaran, S. Permeability barrier of Gram-negative cell
envelopes and approaches to bypass it. _ACS Infect. Dis._ 1, 512–522 (2015). Article CAS PubMed PubMed Central Google Scholar * Zhou, X. et al. Marthiapeptide A, an anti-infective and
cytotoxic polythiazole cyclopeptide from a 60 L scale fermentation of the deep sea-derived _Marinactinospora thermotolerans_ SCSIO 00652. _J. Nat. Prod._ 75, 2251–2255 (2012). Article CAS
PubMed Google Scholar * Grubbs, K. J. et al. Large-scale bioinformatics analysis of _Bacillus_ genomes uncovers conserved roles of natural products in bacterial physiology. _mSystems_ 2,
e00040-17 (2017). Article PubMed PubMed Central Google Scholar * Donia, M. S. et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family
of antibiotics. _Cell_ 158, 1402–1414 (2014). Article CAS PubMed PubMed Central Google Scholar * Russel, J., Røder, H. L., Madsen, J. S., Burmølle, M. & Sørensen, S. J. Antagonism
correlates with metabolic similarity in diverse bacteria. _Proc. Natl Acad. Sci. USA_ 114, 10684–10688 (2017). Article CAS PubMed PubMed Central Google Scholar * Maida, I. et al.
Antagonistic interactions between endophytic cultivable bacterial communities isolated from the medicinal plant _Echinacea purpurea_. _Environ. Microbiol._ 18, 2357–2365 (2016). Article CAS
PubMed Google Scholar * Hassani, M. A., Durán, P. & Hacquard, S. Microbial interactions within the plant holobiont. _Microbiome_ 6, 58 (2018). Article PubMed PubMed Central Google
Scholar * Venturi, V. & Keel, C. Signaling in the rhizosphere. _Trends Plant Sci._ 21, 187–198 (2016). Article CAS PubMed Google Scholar * Chodkowski, J. L. & Shade, A. A
synthetic community system for probing microbial interactions driven by exometabolites. _mSystems_ 2, e00129-17 (2017). Article PubMed PubMed Central Google Scholar * Stringlis, I. A.,
Zhang, H., Pieterse, C. M. J., Bolton, M. D. & de Jonge, R. Microbial small molecules—weapons of plant subversion. _Nat. Prod. Rep._ 35, 410–433 (2018). Article CAS PubMed Google
Scholar * Raaijmakers, J. M. & Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. _Annu. Rev. Phytopathol._ 50, 403–424
(2012). Article CAS PubMed Google Scholar * Peyraud, R. et al. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. _Proc. Natl Acad. Sci. USA_ 106, 4846–4851 (2009).
Article CAS PubMed PubMed Central Google Scholar * Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C. & Segata, N. Compact graphical representation of phylogenetic data and
metadata with GraPhlAn. _PeerJ_ 3, e1029 (2015). Article PubMed PubMed Central Google Scholar * Müller, D. B., Schubert, O. T., Rost, H., Aebersold, R. & Vorholt, J. A. Systems-level
proteomics of two ubiquitous leaf commensals reveals complementary adaptive traits for phyllosphere colonization. _Mol. Cell. Proteom._ 15, 3256–3269 (2016). Article CAS Google Scholar *
Yamanaka, K. et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. _Proc. Natl Acad. Sci. USA_ 111, 1957–1962 (2014).
Article CAS PubMed PubMed Central Google Scholar * Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. _Nucleic Acids Res._ 44, 279–285 (2016).
Article CAS Google Scholar * Ceniceros, A., Dijkhuizen, L., Petrusma, M. & Medema, M. H. Genome-based exploration of the specialized metabolic capacities of the genus _Rhodococcus_.
_BMC Genom._ 18, 593 (2017). Article CAS Google Scholar * Yang, J. Y. et al. Primer on agar-based microbial imaging mass spectrometry. _J. Bacteriol._ 194, 6023–6028 (2012). Article CAS
PubMed PubMed Central Google Scholar * Ueoka, R. et al. Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms. _Nat. Chem. Biol._ 11, 705–712 (2015).
Article CAS PubMed PubMed Central Google Scholar * Durante-Rodríguez, G., de Lorenzo, V. & Martínez-García, E. The Standard European Vector Architecture (SEVA) plasmid toolkit.
_Methods Mol. Biol._ 1149, 469–478 (2014). Article CAS PubMed Google Scholar * Radeck, J. et al. The _Bacillus_ BioBrick Box: generation and evaluation of essential genetic building
blocks for standardized work with _Bacillus subtilis_. _J. Biol. Eng._ 7, 29 (2013). Article PubMed PubMed Central CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was
financially supported by SNF grant NRP72 to J.P. and J.A.V. and by European Research Council Advanced Grants (PhyMo to J.A.V. and SynPlex to J.P.). AUTHOR INFORMATION Author notes * These
authors contributed equally: Eric J. N. Helfrich, Christine M. Vogel. AUTHORS AND AFFILIATIONS * Institute of Microbiology, ETH Zurich, Zurich, Switzerland Eric J. N. Helfrich, Christine M.
Vogel, Reiko Ueoka, Martin Schäfer, Florian Ryffel, Daniel B. Müller, Silke Probst, Markus Kreuzer, Jörn Piel & Julia A. Vorholt Authors * Eric J. N. Helfrich View author publications
You can also search for this author inPubMed Google Scholar * Christine M. Vogel View author publications You can also search for this author inPubMed Google Scholar * Reiko Ueoka View
author publications You can also search for this author inPubMed Google Scholar * Martin Schäfer View author publications You can also search for this author inPubMed Google Scholar *
Florian Ryffel View author publications You can also search for this author inPubMed Google Scholar * Daniel B. Müller View author publications You can also search for this author inPubMed
Google Scholar * Silke Probst View author publications You can also search for this author inPubMed Google Scholar * Markus Kreuzer View author publications You can also search for this
author inPubMed Google Scholar * Jörn Piel View author publications You can also search for this author inPubMed Google Scholar * Julia A. Vorholt View author publications You can also
search for this author inPubMed Google Scholar CONTRIBUTIONS E.J.N.H., C.M.V., R.U., M.S., F.R., D.B.M., J.P. and J.A.V. designed the research. C.M.V., M.S., F.R., D.B.M. and M.K. performed
binary interaction screens. E.J.N.H., C.M.V., F.R. and S.P. performed genome mining studies. C.M.V. and D.B.M. conducted statistical analyses. E.J.N.H., C.M.V. and M.S. conducted MALDI
imaging experiments. E.J.N.H., C.M.V., M.S., F.R. and S.P. conducted bioassays. E.J.N.H., C.M.V., R.U., F.R. and S.P. isolated and structure-elucidated metabolites. M.S. generated
_Brevibacillus_ knockout mutants. E.J.N.H., C.M.V., D.B.M., J.P. and J.A.V. wrote the manuscript with contributions from all authors. CORRESPONDING AUTHORS Correspondence to Jörn Piel or
Julia A. Vorholt. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figures 1–48, Supplementary Results 1, Supplementary
Methods, Supplementary References. REPORTING SUMMARY SUPPLEMENTARY TABLE Supplementary Tables 1–26. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Helfrich, E.J.N., Vogel, C.M., Ueoka, R. _et al._ Bipartite interactions, antibiotic production and biosynthetic potential of the _Arabidopsis_ leaf microbiome. _Nat Microbiol_ 3, 909–919
(2018). https://doi.org/10.1038/s41564-018-0200-0 Download citation * Received: 22 September 2017 * Accepted: 18 June 2018 * Published: 23 July 2018 * Issue Date: August 2018 * DOI:
https://doi.org/10.1038/s41564-018-0200-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
