Global phylogeography and ancient evolution of the widespread human gut virus crassphage
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Microbiomes are vast communities of microorganisms and viruses that populate all natural ecosystems. Viruses have been considered to be the most variable component of microbiomes,
as supported by virome surveys and examples of high genomic mosaicism. However, recent evidence suggests that the human gut virome is remarkably stable compared with that of other
environments. Here, we investigate the origin, evolution and epidemiology of crAssphage, a widespread human gut virus. Through a global collaboration, we obtained DNA sequences of crAssphage
from more than one-third of the world’s countries and showed that the phylogeography of crAssphage is locally clustered within countries, cities and individuals. We also found fully
colinear crAssphage-like genomes in both Old-World and New-World primates, suggesting that the association of crAssphage with primates may be millions of years old. Finally, by exploiting a
large cohort of more than 1,000 individuals, we tested whether crAssphage is associated with bacterial taxonomic groups of the gut microbiome, diverse human health parameters and a wide
range of dietary factors. We identified strong correlations with different clades of bacteria that are related to Bacteroidetes and weak associations with several diet categories, but no
significant association with health or disease. We conclude that crAssphage is a benign cosmopolitan virus that may have coevolved with the human lineage and is an integral part of the
normal human gut virome. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal
Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy
now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS PHYLOGENY AND DISEASE ASSOCIATIONS OF A WIDESPREAD AND ANCIENT INTESTINAL BACTERIOPHAGE LINEAGE Article Open access 27 July 2024 METAGENOMIC
COMPENDIUM OF 189,680 DNA VIRUSES FROM THE HUMAN GUT MICROBIOME Article Open access 24 June 2021 METAGENOMIC ANALYSIS REVEALS UNEXPLORED DIVERSITY OF ARCHAEAL VIROME IN THE HUMAN GUT Article
Open access 29 December 2022 DATA AVAILABILITY Sequence data that support the findings of this study have been deposited in GenBank under BioProject accession PRJNA510571 and at
https://github.com/linsalrob/crAssphage. Each of the samples has a unique BioSample accession number (SAMN10656826–SAMN10658627, SAMN10658653 and SAMN10659294). The SRA runs used in this
analysis are included in Supplementary File 5. The data that support the findings of this study are also available from the corresponding authors on reasonable request. CODE AVAILABILITY The
code used to generate the data can be accessed at https://github.com/linsalrob/crAssphage. The current release81 is v.2.0. REFERENCES * Sender, R., Fuchs, S. & Milo, R. Are we really
vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. _Cell_ 164, 337–340 (2016). Article CAS Google Scholar * Nguyen, S. et al. Bacteriophage transcytosis
provides a mechanism to cross epithelial cell layers. _mBio_ 8, e01874–17 (2017). CAS PubMed PubMed Central Google Scholar * Reyes, A. et al. Viruses in the faecal microbiota of
monozygotic twins and their mothers. _Nature_ 466, 334–338 (2010). Article CAS Google Scholar * Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to
diet. _Genome Res._ 21, 1616–1625 (2011). Article CAS Google Scholar * Reyes, A., Semenkovich, N. P., Whiteson, K., Rohwer, F. & Gordon, J. I. Going viral: next-generation sequencing
applied to phage populations in the human gut. _Nat. Rev. Microbiol._ 10, 607–617 (2012). Article CAS Google Scholar * Paterson, S. et al. Antagonistic coevolution accelerates molecular
evolution. _Nature_ 464, 275–278 (2010). Article CAS Google Scholar * Pedulla, M. L. et al. Origins of highly mosaic mycobacteriophage genomes. _Cell_ 113, 171–182 (2003). Article CAS
Google Scholar * Heldal, M. & Bratbak, G. Production and decay of viruses in aquatic environments. _Mar. Ecol. Prog. Ser._ 72, 205–212 (1991). Article Google Scholar * Breitbart, M.,
Wegley, L., Leeds, S., Schoenfeld, T. & Rohwer, F. Phage community dynamics in hot springs. _Appl. Environ. Microbiol._ 70, 1633–1640 (2004). Article CAS Google Scholar * Steward, G.
F., Smith, D. C. & Azam, F. Abundance and production of bacteria and viruses in the Bering and Chukchi Seas. _Mar. Ecol. Prog. Ser._ 131, 287–300 (1996). Article Google Scholar *
Minot, S. et al. Rapid evolution of the human gut virome. _Proc. Natl Acad. Sci. USA_ 110, 12450–12455 (2013). Article CAS Google Scholar * Dutilh, B. E. et al. A highly abundant
bacteriophage discovered in the unknown sequences of human faecal metagenomes. _Nat. Commun._ 5, 4498 (2014). Article CAS Google Scholar * Yutin, N. et al. Discovery of an expansive
bacteriophage family that includes the most abundant viruses from the human gut. _Nat. Microbiol._ 3, 38–46 (2018). Article CAS Google Scholar * Shkoporov, A. et al. ΦCrAss001, a member
of the most abundant bacteriophage family in the human gut, infects Bacteroides. Preprint at https://doi.org/10.1101/354837 (2018). * Barylski, J. et al. Analysis of spounaviruses as a case
study for the overdue reclassification of tailed bacteriophages. Preprint at https://doi.org/10.1101/220434 (2018). * Adriaenssens, E. & Brister, J. R. How to name and classify your
phage: an informal guide. _Viruses_ 9, 70 (2017). Article Google Scholar * Callahan, B. J., McMurdie, P. J. & Holmes, S. P. Exact sequence variants should replace operational taxonomic
units in marker-gene data analysis. _ISME J._ 11, 2639–2643 (2017). Article Google Scholar * NCBI Resource Coordinators Database resources of the National Center for Biotechnology
Information. _Nucleic Acids Res._ 44, D7–D19 (2016). Article Google Scholar * Nicholls, S. M. et al. Probabilistic recovery of cryptic haplotypes from metagenomic data. Preprint at
https://doi.org/10.1101/117838 (2017). * Lim, E. S. et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. _Nat. Med._ 21, 1228–1234 (2015). Article CAS
Google Scholar * Liang, Y. Y., Zhang, W., Tong, Y. G. & Chen, S. P. crAssphage is not associated with diarrhoea and has high genetic diversity. _Epidemiol. Infect._ 144, 3549–3553
(2016). Article CAS Google Scholar * Piper, H. G. et al. Severe gut microbiota dysbiosis is associated with poor growth in patients with short bowel syndrome. _JPEN J. Parenter. Enter.
Nutr._ 41, 1202–1212 (2017). Article Google Scholar * Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. _Cell_ 165, 842–853 (2016).
Article CAS Google Scholar * Huerta-Cepas, J., Serra, F. & Bork, P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. _Mol. Biol. Evol._ 33, 1635–1638 (2016).
Article CAS Google Scholar * Stachler, E. et al. Quantitative crAssphage PCR assays for human fecal pollution measurement. _Environ. Sci. Technol._ 51, 9146–9154 (2017). Article CAS
Google Scholar * Stachler, E. & Bibby, K. Metagenomic evaluation of the highly abundant human gut bacteriophage crAssphage for source tracking of human fecal pollution. _Environ. Sci.
Technol. Lett._ 1, 405–409 (2014). Article CAS Google Scholar * García-Aljaro, C., Ballesté, E., Muniesa, M. & Jofre, J. Determination of crAssphage in water samples and applicability
for tracking human faecal pollution. _Microb. Biotechnol._ 10, 1775–1780 (2017). Article Google Scholar * Ahmed, W. et al. Evaluation of the novel crAssphage marker for sewage pollution
tracking in storm drain outfalls in Tampa, Florida. _Water Res._ 131, 142–150 (2017). Article Google Scholar * Yatsunenko, T. et al. Human gut microbiome viewed across age and geography.
_Nature_ 486, 222–227 (2012). Article CAS Google Scholar * Santiago-Rodriguez, T. M. et al. Natural mummification of the human gut preserves bacteriophage DNA. _FEMS Microbiol. Lett._
363, fnv219 (2016). Article Google Scholar * Maixner, F. et al. The 5300-year-old _Helicobacter pylori_ genome of the Iceman. _Science_ 351, 162–165 (2016). Article CAS Google Scholar *
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. _J. Mol. Biol._ 215, 403–410 (1990). Article CAS Google Scholar * Guerin, E. et
al. Biology and taxonomy of crAss-like bacteriophages, the most abundant virus in the human gut. _Cell Host Microbe_ 24, 653–664 (2018). Article CAS Google Scholar * Raymond, F. et al.
The initial state of the human gut microbiome determines its reshaping by antibiotics. _ISME J._ 10, 707–720 (2016). Article CAS Google Scholar * Moeller, A. H. et al. Rapid changes in
the gut microbiome during human evolution. _Proc. Natl Acad. Sci. USA_ 111, 16431–16435 (2014). Article CAS Google Scholar * Moeller, A. H. et al. Cospeciation of gut microbiota with
hominids. _Science_ 353, 380–382 (2016). Article CAS Google Scholar * Zhernakova, A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and
diversity. _Science_ 352, 565–569 (2016). Article CAS Google Scholar * Tigchelaar, E. F. et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the
northern Netherlands: study design and baseline characteristics. _BMJ Open_ 5, e006772 (2015). Article Google Scholar * David, L. A. et al. Diet rapidly and reproducibly alters the human
gut microbiome. _Nature_ 505, 559–563 (2014). Article CAS Google Scholar * Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized
gnotobiotic mice. _Sci. Transl. Med._ 1, 6ra14 (2009). Article Google Scholar * Singh, R. K. et al. Influence of diet on the gut microbiome and implications for human health. _J. Transl.
Med._ 15, 73 (2017). Article Google Scholar * De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa.
_Proc. Natl Acad. Sci. USA_ 107, 14691–14696 (2010). Article Google Scholar * Kovatcheva-Datchary, P. et al. Dietary fiber-induced improvement in glucose metabolism is associated with
increased abundance of _Prevotella_. _Cell Metab._ 22, 971–982 (2015). Article CAS Google Scholar * Edwards, R. A., McNair, K., Faust, K., Raes, J. & Dutilh, B. E. Computational
approaches to predict bacteriophage-host relationships. _FEMS Microbiol. Rev._ 40, 258–272 (2016). Article CAS Google Scholar * Manrique, P. et al. Healthy human gut phageome. _Proc. Natl
Acad. Sci. USA_ 113, 10400–10405 (2016). Article CAS Google Scholar * Kupczok, A. et al. Rates of mutation and recombination in _Siphoviridae_ phage genome evolution over three decades.
_Mol. Biol. Evol._ 35, 1147–1159 (2018). Article CAS Google Scholar * Schrago, C. G. & Russo, C. A. M. Timing the origin of New World monkeys. _Mol. Biol. Evol._ 20, 1620–1625 (2003).
Article CAS Google Scholar * Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. _J. Comput. Biol._ 19, 455–477 (2012). Article
CAS Google Scholar * Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. _BMC Bioinform._ 11, 119 (2010). Article Google Scholar *
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. _Mol. Syst. Biol._ 7, 539 (2011). Article Google Scholar * Nguyen,
L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. _Mol. Biol. Evol._ 32, 268–274
(2015). Article CAS Google Scholar * Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic
estimates. _Nat. Methods_ 14, 587–589 (2017). Article CAS Google Scholar * Zhou, X., Shen, X., Hittinger, C. T. & Rokas, A. Evaluating fast maximum likelihood-based phylogenetic
programs using empirical phylogenomic data sets. _Mol. Biol. Evol._ 35, 486–503 (2017). Article Google Scholar * Dutilh, B. E. et al. Assessment of phylogenomic and orthology approaches
for phylogenetic inference. _Bioinformatics_ 23, 815–824 (2007). Article CAS Google Scholar * Cinek, O. et al. Quantitative crAssphage real-time PCR assay derived from data of multiple
geographically distant populations. _J. Med. Virol._ 90, 767–771 (2018). Article CAS Google Scholar * Liang, Y., Jin, X., Huang, Y. & Chen, S. Development and application of a
real-time polymerase chain reaction assay for detection of a novel gut bacteriophage (crAssphage). _J. Med. Virol._ 90, 464–468 (2018). Article CAS Google Scholar * Langmead, B. &
Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. _Nat. Methods_ 9, 357–359 (2012). Article CAS Google Scholar * Li, H. et al. The Sequence Alignment/Map format and SAMtools.
_Bioinformatics_ 25, 2078–2079 (2009). Article Google Scholar * Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. _Genome Res._ 8,
186–194 1998). Article CAS Google Scholar * Ewing, B., Hillier, L., Wendl, M. C. & Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. _Genome
Res._ 8, 175–185 (1998). Article CAS Google Scholar * Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. _Trends Genet._ 16, 276–277
(2000). Article CAS Google Scholar * Knudsen, B. E., Bergmark, L. & Pamp, S. J. SOP—DNA isolation QIAamp Fast DNA Stool modified. _Figshare_
https://doi.org/10.6084/m9.figshare.3475406.v4 (2016). * National Center for Biotechnology Information SRA Handbook (National Center for Biotechnology Information, 2009). * Stewart, C. A. et
al. Jetstream: a self-provisioned, scalable science and engineering cloud environment. In _Proc. 2015 XSEDE Conference Scientific Advancements Enabled by Enhanced Cyberinfrastructure_ 29
(ACM, 2015). * Towns, J. et al. XSEDE: accelerating scientific discovery. _Comput. Sci. Eng._ 16, 62–74 (2014). Article Google Scholar * Edwards, R. SearchSRA (2017);
https://doi.org/10.5281/zenodo.1043562 * Torres, P. J., Edwards, R. A. & McNair, K. PARTIE: a partition engine to separate metagenomics and amplicon projects in the Sequence Read
Archive. _Bioinformatics_ 33, 2389–2391 (2017). Article CAS Google Scholar * Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. _Bioinformatics_
27, 863–864 (2011). Article CAS Google Scholar * Cantu, V. A., Sadural, J. & Edwards, R. PRINSEQ++, a multi-threaded tool for fast and efficient quality control and preprocessing of
sequencing datasets. Preprint at https://doi.org/10.7287/peerj.preprints.27553v1 (2019). * Levi, K., Rynge, M., Eroma, A. & Edwards, R. A. Searching the Sequence Read Archive using
Jetstream and Wrangler. In _Proc. Practice and Experience on Advanced Research Computing_ (2018)_._ * Stallman, R. M., McGrath, R. & Smith, P. D. _GNU Make: A Program for Directing
Recompilation, for version 3.81_ (Free Software Foundation, 2004). * Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. _Nucleic Acids Res._ 32,
1792–1797 (2004). Article CAS Google Scholar * Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other
trees. _Nucleic Acids Res._ 44, W242–W245 (2016). Article CAS Google Scholar * Berke, L. & Snel, B. The histone modification H3K27me3 is retained after gene duplication and correlates
with conserved noncoding sequences in _Arabidopsis_. _Genome Biol. Evol._ 6, 572–579 (2014). Article CAS Google Scholar * Meyer, F. et al. The metagenomics RAST server—a public resource
for the automatic phylogenetic and functional analysis of metagenomes. _BMC Bioinform._ 9, 386 (2008). Article CAS Google Scholar * Zhao, Y., Tang, H. & Ye, Y. RAPSearch2: a fast and
memory-efficient protein similarity search tool for next-generation sequencing data. _Bioinformatics_ 28, 125–126 (2012). Article CAS Google Scholar * Vlčková, K. et al. Impact of stress
on the gut microbiome of free-ranging western lowland gorillas. _Microbiology_ 164, 40–44 (2018). Article Google Scholar * McNair, K., Zhou, C., Dinsdale, E. A., Souza, B. & Edwards,
R. A. PHANOTATE: a novel approach to gene identification in phage genomes. _Bioinformatics_ https://doi.org/10.1093/bioinformatics/btz265 (2019). * Katoh, K. & Standley, D. M. MAFFT
multiple sequence alignment software version 7: improvements in performance and usability. _Mol. Biol. Evol._ 30, 772–780 (2013). Article CAS Google Scholar * Benjamini, Y. &
Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. _J. R. Stat. Soc. Series B Stat. Methodol._ 57, 289–300 (1995). Google Scholar *
Dutilh, Bas E., and Edwards, Robert A. _crAssphage Data Repository on GitHu_b (Github, 2018); https://doi.org/10.5281/zenodo.1230436 Download references ACKNOWLEDGEMENTS We thank R.
Matthews, M. Wright, J. Alexander, S. Arredondo, N. Branch, D. Campbell, R. Chea, D. McDougle, J. Parks and V. Vipatapat for providing access to wastewater treatment samples; the members of
the Mountain Gorilla Veterinary Project and the staff of Maryland Zoo for collecting the gorilla faecal samples in Rwanda; G. Britton for collecting the baboon faecal samples in Ethiopia;
staff of the CSWCT, the UWA and the UNCST for collecting the chimpanzee faecal samples in Uganda; J. Manor at Central Virology Laboratory, Chaim Sheba Medical Center, Tel-Hashomer Hospital
and G. Steward, Department of Oceanography, University of Hawai’i at Manoa for help with sample collection; the COMPARE and LifeLines-DEEP projects for sharing data; O.D.N. thanks G.
Steward, University of Hawai’i, Manoa for support. P.C.F. thanks C. Taylor for support with the PCR. Primate samples were provided by the PMC at the University of Illinois Urbana-Champaign;
D.T.M. thanks the Australian Research Council’s Linkage Project LP160100408, Melbourne Water and EPA Victoria for funding the collection of samples in Melbourne. Gorilla samples were
originally obtained by M.K. and the Mountain Gorilla Veterinary Project in Rwanda. G.R. and N.J.D. provided the wild baboon samples from Ethiopia. Howler samples were provided by M.K. and
lemur samples were provided by R.E.J. and M.T.I., R.M.S. and L.M. provided the chimpanzee samples with permission from the CSWCT, the UWA and the UNCST. The primate microbiome project was
supported by NSF BCS 0935347 to S.L., R. Stumpf, B.W. and K. Nelson. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the
manuscript. This work used the XSEDE Jetstream resources at Indiana University and Texas Advanced Computing Center through allocation MCB170036 to R.A.E., which is supported by National
Science Foundation grant number ACI-1548562. Some of this work was supported by San Diego State University Grants Programs to R.A.E., including the Summer Undergraduate Research Program.
This work was supported by National Science Foundation grant numbers MCB-1441985 to R.A.E. and DUE-1323809 to E.A.D; the Department of Energy Lawrence Livermore National Laboratory grant
B618146 to R.A.E., P.A.d.J. and B.E.D. were supported by the NWO Vidi grant 864.14.004; F.L.N. by the NWO Veni grant 016.Veni.181.092; S.J.J.B. by the European Research Council Stg grant
(638707) and the Vidi grant 864.11.005; O.C. and K. Mazankova by the Ministry of Health of the Czech Republic grant numbers 15-31426A and 15-29078A; P.C.F. by a Rutherford Discovery
Fellowship from the Royal Society of New Zealand. J.J.B. by the ARC Discovery Early Career Researcher Award (DE170100525); S.L.D.M. by an NIH Pathway to Independence Fellowship
(1K99AI119401-01A1); K.B. by award number 1510925 from the United States National Science Foundation; M.T.I. by National Geographic Society (CRE) and NSERC; and C.D. by the Agence Nationale
de la Recherche JCJC grant ANR-13-JSV6-0004 and Investissements d’Avenir Méditerranée Infection 10-IAHU-03. The LifeLines-DEEP sample collection and analysis was funded by the Netherlands
Heart Foundation (IN-CONTROL CVON grant 2012-03) to A.Z. and J.F., by the Top Institute Food and Nutrition, Wageningen, the Netherlands (TiFN GH001) to C.W., by NWO Vidi grants 864.13.013 to
J.F. and 016.178.056 to A.Z., NWO Spinoza Prize SPI 92-266 to C.W., and by the ERC FP7/2007-2013/ERC Advanced Grant agreement 2012-322698 to C.W., ERC Starting Grant 715772 to A.Z. A.Z.
also holds a Rosalind Franklin Fellowship from the University of Groningen. The COMPARE data collection was funded by The Novo Nordisk Foundation (NNF16OC0021856). AUTHOR INFORMATION AUTHORS
AND AFFILIATIONS * Department of Biology, San Diego State University, San Diego, CA, USA Robert A. Edwards, Alejandro A. Vega, Holly M. Norman, Maria Ohaeri, Elizabeth A. Dinsdale, Emma K.
Billings, Tess Condeff, Michael P. Doane, John M. Haggerty, Scott T. Kelley, David Lipson, Megan Morris, Kim Reasor & Pedro J. Torres * The Viral Information Institute, San Diego State
University, San Diego, CA, USA Robert A. Edwards * Department of Computer Science, San Diego State University, San Diego, CA, USA Kyle Levi * Department of Pediatrics, 2nd Faculty of
Medicine, Charles University in Prague, Prague, Czech Republic Ondrej Cinek & Karla Mazankova * Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo,
Egypt Ramy K. Aziz * Computational Sciences Research Center, San Diego State University, San Diego, CA, USA Katelyn McNair, Vito Adrian Cantu & Daniel A. Cuevas * School of Biological
Sciences, Monash University, Clayton, Victoria, Australia Jeremy J. Barr * Civil and Environmental Engineering and Earth Sciences, University of Notre Dame, Notre Dame, IN, USA Kyle Bibby *
Department of Bionanoscience, Kavli Institute of Nanoscience, Delft University of Technology, Delft, The Netherlands Stan J. J. Brouns, Patrick A. de Jonge & Franklin L. Nobrega *
Institute of Infection and Global Health, University of Liverpool, Liverpool, UK Adrian Cazares * Theoretical Biology and Bioinformatics, Science4Life, Utrecht University, Utrecht, The
Netherlands Patrick A. de Jonge, Alessandro Rossi & Bas E. Dutilh * MEPHI, Aix-Marseille Université, IRD, AP-HM, CNRS, IHU Méditerranée Infection, Marseille, France Christelle Desnues *
Mediterranean Institute of Oceanography, Aix-Marseille Université, Université de Toulon, CNRS, IRD, UM 110, Marseille, France Christelle Desnues * Center for Genomics and Systems Biology
& Department of Biology, New York University, New York, NY, USA Samuel L. Díaz Muñoz, Jane M. Carlton, Elodie Ghedin, Kristen M. Gulino, Julia M. Maritz & Alan Twaddle * Department
of Microbiology and Molecular Genetics, University of California, Davis, Davis, CA, USA Samuel L. Díaz Muñoz * Department of Microbiology and Immunology, University of Otago, Dunedin, New
Zealand Peter C. Fineran * Department of Genetics, University Medical Center Groningen, Groningen, The Netherlands Alexander Kurilshikov, Alexandra Zhernakova & Cisca Wijmenga *
Department of Biosystems, KU Leuven, Leuven, Belgium Rob Lavigne & Jeroen Wagemans * EPHM Lab, Civil Engineering Department, Monash University, Clayton, Victoria, Australia David T.
McCarthy * Max Planck Tandem Group in Computational Biology, Departamento de Ciencias Biológicas, Universidad de los Andes, Bogotá, Colombia Alejandro Reyes Muñoz * Department of Child
Health, Norwegian Institute of Public Health, Oslo, Norway German Tapia & Lars C. Stene * GEMA Center for Genomics, Ecology & Environment, Universidad Mayor, Huechuraba, Chile Nicole
Trefault * Laboratory of Bioinformatics, Federal Research and Clinical Center of Physical-Chemical Medicine, Moscow, Russia Alexander V. Tyakht * Department of Informational Technologies,
ITMO University, Saint Petersburg, Russia Alexander V. Tyakht * Centro de Ciencias Genómicas, Universidad Nacional Autónoma de México, Cuernavaca, Mexico Pablo Vinuesa & Daniel Cazares *
National Food Institute, Research Group for Genomic Epidemiology, Technical University of Denmark, Kongens Lyngby, Denmark Frank M. Aarestrup & Rene S. Hendriksen * Endocrine Centre
Baku, Baku, Azerbaijan Gunduz Ahmadov * Department of Pediatrics, School of Medicine, University of Jordan, Amman, Jordan Abeer Alassaf & Rasha Odeh * Department of Physiology, Genetics
and Microbiology, University of Alicante, Alicante, Spain Josefa Anton * Carl R. Woese Institute of Genomic Biology, University of Illinois at Urbana-Champaign, Urbana, IL, USA Abigail
Asangba, Rebecca Stumpf & Bryan White * Department of Microbiology and Biotechnology, Max Rubner-Institut, Federal Research Institute of Nutrition and Food, Kiel, Germany Gyu-Sung Cho,
Charles Franz & Horst Neve * Departament de Genètica i de Microbiologia, Universitat Autònoma De Barcelona, Barcelona, Spain Pilar Cortés & Montserrat Llagostera * Wildlife Health
Center, University of California, Davis, Davis, CA, USA Mike Cranfield * Departamento de Genética Molecular y Microbiología, Pontificia Universidad Católica de Chile, Santiago, Chile Rodrigo
De la Iglesia * Department of Bacterial Genetics, Institute of Microbiology, Faculty of Biology, University of Warsaw, Warsaw, Poland Przemyslaw Decewicz & Lukasz Dziewit * Department
of Anthropology, Dartmouth College, Hanover, NH, USA Nathaniel J. Dominy & Gillian A. O. Rice * Department of Pediatrics and Child Health, Faculty of Medicine, University of Khartoum,
Khartoum, Sudan Bashir Mukhtar Elwasila * Department of Medicine, University of Chicago, Chicago, IL, USA A. Murat Eren * Department of Pediatrics, University Medical Center Groningen,
Groningen, The Netherlands Jingyuan Fu * Department of Genetics, Microbiology and Statistics, Universitat de Barcelona, Barcelona, Spain Cristina Garcia-Aljaro, Juan Jofre & Maite
Muniesa * Next Generation Sequencing and Microarray Core Facility, The Scripps Research Institute, La Jolla, CA, USA Steven R. Head * School of Microbiology, University College Cork, Cork,
Ireland Colin Hill * Department of Virology, School of Medicine, University of Tampere, Tampere, Finland Heikki Hyöty * Department of Molecular Biology and Genetics, Federal Research and
Clinical Center of Physical-Chemical Medicine, Moscow, Russia Elena N. Ilina * Department of Anthropology, Northern Illinois University, DeKalb, IL, USA Mitchell T. Irwin * School of Science
and Health, Western Sydney University, Penrith, New South Wales, Australia Thomas C. Jeffries * Department of Animal Health, Columbus Zoo and Aquarium, Powell, OH, USA Randall E. Junge *
Department of Microbiology and Immunology, McGill University, Montreal, Quebec, Canada Mohammadali Khan Mirzaei * Department Estacion Biologica Corrientes, Institution Museo Arg. Cs.
Naturales-CONICET, Corrientes, Argentina Martin Kowalewski * UWA School of Agriculture and Environment, University of Western Australia, Perth, Western Australia, Australia Deepak Kumaresan,
Benjamin Moreira-Grez & Andy Whiteley * Department of Anthropology, University of Colorado, Boulder, CO, USA Steven R. Leigh * Department of Research and Development, Lytech Ltd.,
Moscow, Russia Eugenia S. Lisitsyna * Department of Civil and Environmental Engineering, Virginia Tech, Blacksburg, VA, USA Linsey C. Marr, Aaron J. Prussin II & John Shimashita * APC
Microbiome Institute, University College Cork, Cork, Ireland Angela McCann & Ronan Strain * Clinical Microbiology & Immunology, Sackler school of Medicine, Tel Aviv University, Tel
Aviv, Israel Shahar Molshanski-Mor * Laboratorio de Analises, Instituto Superior Tecnico, Universidade Lisboa, Lisboa, Portugal Silvia Monteiro & Ricardo Santos * CEHA, Kampala, Uganda
Lawrence Mugisha * COVAB, Makerere University, Kampala, Uganda Lawrence Mugisha * Computer Science and Engineering, University of California, San Diego, La Jolla, CA, USA Nam-phuong Nguyen *
College of Natural and Computational Sciences, Hawai’i Pacific University, Kaneohe, HI, USA Olivia D. Nigro * Department of Molecular Biosciences, Stockholm University, Stockholm, Sweden
Anders S. Nilsson * Biological and Medical Informatics Program, San Diego State University, San Diego, CA, USA Taylor O’Connell * Department of Molecular Biology & Biochemistry,
University of California, Irvine, Irvine, CA, USA Andrew Oliver, Stephen Wandro & Katrine L. Whiteson * Departamento de Química Biológica, Facultad de Ciencias Exactas y Naturales,
Universidad de Buenos Aires, Buenos Aires, Argentina Mariana Piuri * Department of Clinical Microbiology and Immunology, Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel Udi
Qimron * Ministry of Education Key Laboratory for Biodiversity Science and Ecological Engineering, Fudan University, Shanghai, China Zhe-Xue Quan * Centre of Epidemiology and Microbiology,
National Institute of Public Health, Prague, Czech Republic Petra Rainetova * Molecular Genetics, Corporación Corpogen, Bogotá, Colombia Adán Ramírez-Rojas & Maria M. Zambrano * CERELA,
Tucumán, Argentina Raul Raya * Department of Biology, University of Padova, Padova, Italy Alessandro Rossi * Swanson School of Engineering, University of Pittsburgh, Pittsburgh, PA, USA
Elyse N. Stachler * Department of Pediatrics, Federal Teaching Hospital Abakaliki, Ebonyi State University, Abakaliki, Nigeria MaryAnn Ugochi Ibekwe * Escuela de Tecnología Médica,
Universidad Andres Bello, Santiago, Chile Nicolás Villagra * The Bioinformatics Centre, Department of Biology, University of Copenhagen, Copenhagen, Denmark Henrike Zschach * Centre for
Molecular and Biomolecular Informatics, Radboud Institute for Molecular Life Sciences, Radboud University Medical Centre, Nijmegen, The Netherlands Bas E. Dutilh Authors * Robert A. Edwards
View author publications You can also search for this author inPubMed Google Scholar * Alejandro A. Vega View author publications You can also search for this author inPubMed Google Scholar
* Holly M. Norman View author publications You can also search for this author inPubMed Google Scholar * Maria Ohaeri View author publications You can also search for this author inPubMed
Google Scholar * Kyle Levi View author publications You can also search for this author inPubMed Google Scholar * Elizabeth A. Dinsdale View author publications You can also search for this
author inPubMed Google Scholar * Ondrej Cinek View author publications You can also search for this author inPubMed Google Scholar * Ramy K. Aziz View author publications You can also search
for this author inPubMed Google Scholar * Katelyn McNair View author publications You can also search for this author inPubMed Google Scholar * Jeremy J. Barr View author publications You
can also search for this author inPubMed Google Scholar * Kyle Bibby View author publications You can also search for this author inPubMed Google Scholar * Stan J. J. Brouns View author
publications You can also search for this author inPubMed Google Scholar * Adrian Cazares View author publications You can also search for this author inPubMed Google Scholar * Patrick A. de
Jonge View author publications You can also search for this author inPubMed Google Scholar * Christelle Desnues View author publications You can also search for this author inPubMed Google
Scholar * Samuel L. Díaz Muñoz View author publications You can also search for this author inPubMed Google Scholar * Peter C. Fineran View author publications You can also search for this
author inPubMed Google Scholar * Alexander Kurilshikov View author publications You can also search for this author inPubMed Google Scholar * Rob Lavigne View author publications You can
also search for this author inPubMed Google Scholar * Karla Mazankova View author publications You can also search for this author inPubMed Google Scholar * David T. McCarthy View author
publications You can also search for this author inPubMed Google Scholar * Franklin L. Nobrega View author publications You can also search for this author inPubMed Google Scholar *
Alejandro Reyes Muñoz View author publications You can also search for this author inPubMed Google Scholar * German Tapia View author publications You can also search for this author
inPubMed Google Scholar * Nicole Trefault View author publications You can also search for this author inPubMed Google Scholar * Alexander V. Tyakht View author publications You can also
search for this author inPubMed Google Scholar * Pablo Vinuesa View author publications You can also search for this author inPubMed Google Scholar * Jeroen Wagemans View author publications
You can also search for this author inPubMed Google Scholar * Alexandra Zhernakova View author publications You can also search for this author inPubMed Google Scholar * Frank M. Aarestrup
View author publications You can also search for this author inPubMed Google Scholar * Gunduz Ahmadov View author publications You can also search for this author inPubMed Google Scholar *
Abeer Alassaf View author publications You can also search for this author inPubMed Google Scholar * Josefa Anton View author publications You can also search for this author inPubMed Google
Scholar * Abigail Asangba View author publications You can also search for this author inPubMed Google Scholar * Emma K. Billings View author publications You can also search for this
author inPubMed Google Scholar * Vito Adrian Cantu View author publications You can also search for this author inPubMed Google Scholar * Jane M. Carlton View author publications You can
also search for this author inPubMed Google Scholar * Daniel Cazares View author publications You can also search for this author inPubMed Google Scholar * Gyu-Sung Cho View author
publications You can also search for this author inPubMed Google Scholar * Tess Condeff View author publications You can also search for this author inPubMed Google Scholar * Pilar Cortés
View author publications You can also search for this author inPubMed Google Scholar * Mike Cranfield View author publications You can also search for this author inPubMed Google Scholar *
Daniel A. Cuevas View author publications You can also search for this author inPubMed Google Scholar * Rodrigo De la Iglesia View author publications You can also search for this author
inPubMed Google Scholar * Przemyslaw Decewicz View author publications You can also search for this author inPubMed Google Scholar * Michael P. Doane View author publications You can also
search for this author inPubMed Google Scholar * Nathaniel J. Dominy View author publications You can also search for this author inPubMed Google Scholar * Lukasz Dziewit View author
publications You can also search for this author inPubMed Google Scholar * Bashir Mukhtar Elwasila View author publications You can also search for this author inPubMed Google Scholar * A.
Murat Eren View author publications You can also search for this author inPubMed Google Scholar * Charles Franz View author publications You can also search for this author inPubMed Google
Scholar * Jingyuan Fu View author publications You can also search for this author inPubMed Google Scholar * Cristina Garcia-Aljaro View author publications You can also search for this
author inPubMed Google Scholar * Elodie Ghedin View author publications You can also search for this author inPubMed Google Scholar * Kristen M. Gulino View author publications You can also
search for this author inPubMed Google Scholar * John M. Haggerty View author publications You can also search for this author inPubMed Google Scholar * Steven R. Head View author
publications You can also search for this author inPubMed Google Scholar * Rene S. Hendriksen View author publications You can also search for this author inPubMed Google Scholar * Colin
Hill View author publications You can also search for this author inPubMed Google Scholar * Heikki Hyöty View author publications You can also search for this author inPubMed Google Scholar
* Elena N. Ilina View author publications You can also search for this author inPubMed Google Scholar * Mitchell T. Irwin View author publications You can also search for this author
inPubMed Google Scholar * Thomas C. Jeffries View author publications You can also search for this author inPubMed Google Scholar * Juan Jofre View author publications You can also search
for this author inPubMed Google Scholar * Randall E. Junge View author publications You can also search for this author inPubMed Google Scholar * Scott T. Kelley View author publications You
can also search for this author inPubMed Google Scholar * Mohammadali Khan Mirzaei View author publications You can also search for this author inPubMed Google Scholar * Martin Kowalewski
View author publications You can also search for this author inPubMed Google Scholar * Deepak Kumaresan View author publications You can also search for this author inPubMed Google Scholar *
Steven R. Leigh View author publications You can also search for this author inPubMed Google Scholar * David Lipson View author publications You can also search for this author inPubMed
Google Scholar * Eugenia S. Lisitsyna View author publications You can also search for this author inPubMed Google Scholar * Montserrat Llagostera View author publications You can also
search for this author inPubMed Google Scholar * Julia M. Maritz View author publications You can also search for this author inPubMed Google Scholar * Linsey C. Marr View author
publications You can also search for this author inPubMed Google Scholar * Angela McCann View author publications You can also search for this author inPubMed Google Scholar * Shahar
Molshanski-Mor View author publications You can also search for this author inPubMed Google Scholar * Silvia Monteiro View author publications You can also search for this author inPubMed
Google Scholar * Benjamin Moreira-Grez View author publications You can also search for this author inPubMed Google Scholar * Megan Morris View author publications You can also search for
this author inPubMed Google Scholar * Lawrence Mugisha View author publications You can also search for this author inPubMed Google Scholar * Maite Muniesa View author publications You can
also search for this author inPubMed Google Scholar * Horst Neve View author publications You can also search for this author inPubMed Google Scholar * Nam-phuong Nguyen View author
publications You can also search for this author inPubMed Google Scholar * Olivia D. Nigro View author publications You can also search for this author inPubMed Google Scholar * Anders S.
Nilsson View author publications You can also search for this author inPubMed Google Scholar * Taylor O’Connell View author publications You can also search for this author inPubMed Google
Scholar * Rasha Odeh View author publications You can also search for this author inPubMed Google Scholar * Andrew Oliver View author publications You can also search for this author
inPubMed Google Scholar * Mariana Piuri View author publications You can also search for this author inPubMed Google Scholar * Aaron J. Prussin II View author publications You can also
search for this author inPubMed Google Scholar * Udi Qimron View author publications You can also search for this author inPubMed Google Scholar * Zhe-Xue Quan View author publications You
can also search for this author inPubMed Google Scholar * Petra Rainetova View author publications You can also search for this author inPubMed Google Scholar * Adán Ramírez-Rojas View
author publications You can also search for this author inPubMed Google Scholar * Raul Raya View author publications You can also search for this author inPubMed Google Scholar * Kim Reasor
View author publications You can also search for this author inPubMed Google Scholar * Gillian A. O. Rice View author publications You can also search for this author inPubMed Google Scholar
* Alessandro Rossi View author publications You can also search for this author inPubMed Google Scholar * Ricardo Santos View author publications You can also search for this author
inPubMed Google Scholar * John Shimashita View author publications You can also search for this author inPubMed Google Scholar * Elyse N. Stachler View author publications You can also
search for this author inPubMed Google Scholar * Lars C. Stene View author publications You can also search for this author inPubMed Google Scholar * Ronan Strain View author publications
You can also search for this author inPubMed Google Scholar * Rebecca Stumpf View author publications You can also search for this author inPubMed Google Scholar * Pedro J. Torres View
author publications You can also search for this author inPubMed Google Scholar * Alan Twaddle View author publications You can also search for this author inPubMed Google Scholar * MaryAnn
Ugochi Ibekwe View author publications You can also search for this author inPubMed Google Scholar * Nicolás Villagra View author publications You can also search for this author inPubMed
Google Scholar * Stephen Wandro View author publications You can also search for this author inPubMed Google Scholar * Bryan White View author publications You can also search for this
author inPubMed Google Scholar * Andy Whiteley View author publications You can also search for this author inPubMed Google Scholar * Katrine L. Whiteson View author publications You can
also search for this author inPubMed Google Scholar * Cisca Wijmenga View author publications You can also search for this author inPubMed Google Scholar * Maria M. Zambrano View author
publications You can also search for this author inPubMed Google Scholar * Henrike Zschach View author publications You can also search for this author inPubMed Google Scholar * Bas E.
Dutilh View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS B.E.D. and R.A.E. conceived the study, performed the experiments and bioinformatics,
and wrote the paper with input from all authors. A.A.V. performed the volunteer experiments and sampled San Diego wastewater treatment plants. F.L.N., H.M.N., M.O. and P.A.d.J. performed
human volunteer experiments. A.M.E., A.R., A.T., D.A.C., J.M.H., K.L., K.McNair, T.C. and V.A.C. performed bioinformatics analysis. A.A.R.R., A.Alassaf, A.C., A.M., A.O., A.R.M., A.S.N.,
A.W., B.M.-G., B.M.E., C.D., C.F., C.H., D.C., D.K., D.T.M., E.A.D., E.B., E.N.I., E.N.S., E.S.L., G.A., G.C.-A., G.-S.C., G.T., H.H., H.N., J.A., J.J.B., J.J.T., J.M.C., J.M.M., J.W., K.B.,
K.L.W., K.Mazankova, L.C.S., L.D., M.A.U.I., M.K.M., M.L., M.M.Z., M.Morris, M.Muniesa, M.P., M.P.D., N.T., N.V., O.C., O.D.N., P.C., P.C.F., P.D., P.R., P.V., R.d.l.I., R.K.A., R.L., R.O.,
R.R., R.Santos, R.Strain, S.J.J.B., S.L.D.M., S.M., S.M.-M., S.W., T.C., T.J., U.Q. and Z.-X.Q. performed sampling, PCR and sequencing. A.K., A.Z., C.W. and J.F. performed the Lifelines
analysis. F.M.A., H.Z. and R.S.H. provided and analysed COMPARE project data. A.Asangba, B.W., G.A.O.R., N.J.D., N.-p.N., R.Stumpf and S.L. analysed and provided the non-human primate
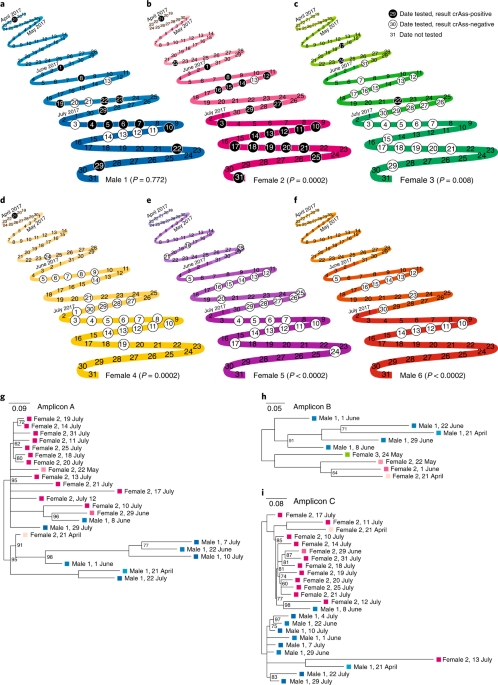
sequences. M.C. collected gorilla samples. A.T., E.G. and K.M.G. performed the NYC sewage sampling and data analysis. A.J.P., J.S., L.C.M., P.J.T., S.R.H. and S.T.K. examined crAssphage
transfer among infants. M.T.I. and R.E.J. collected lemur samples. M.K. collected howler monkey samples. D.L., K.R. created the map of the world figure. L.M. collected chimpanzee samples.
CORRESPONDING AUTHORS Correspondence to Robert A. Edwards or Bas E. Dutilh. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION
PUBLISHER’S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION
Supplementary Figs. 1–9, Supplementary Tables 1–6 and Supplementary References. REPORTING SUMMARY SUPPLEMENTARY FILE 1 Global sampling of crAssphage: the metadata and sequence data for each
of the amplicon regions. SUPPLEMENTARY FILE 2 Gretel strains: number of strains identified from all of the different samples in the SRA. SUPPLEMENTARY FILE 3 Lifelines phenotype
correlations: correlation, _P_ value and adjusted _P_ value for 207 exogenous and intrinsic human variables, and the presence of crAssphage in stools. SUPPLEMENTARY FILE 4 Lifelines
microbial correlations: correlation, _P_ value and adjusted _P_ value for the presence of 491 bacterial isolates and the presence of crAssphage in stools. SUPPLEMENTARY FILE 5 SRA Runs: the
identities of all runs in the SRA with matches to crAssphage, including the number of sequences that match, the total bases aligned and the average coverage. RIGHTS AND PERMISSIONS Reprints
and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Edwards, R.A., Vega, A.A., Norman, H.M. _et al._ Global phylogeography and ancient evolution of the widespread human gut virus
crAssphage. _Nat Microbiol_ 4, 1727–1736 (2019). https://doi.org/10.1038/s41564-019-0494-6 Download citation * Received: 14 October 2018 * Accepted: 22 May 2019 * Published: 08 July 2019 *
Issue Date: October 2019 * DOI: https://doi.org/10.1038/s41564-019-0494-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link
Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
