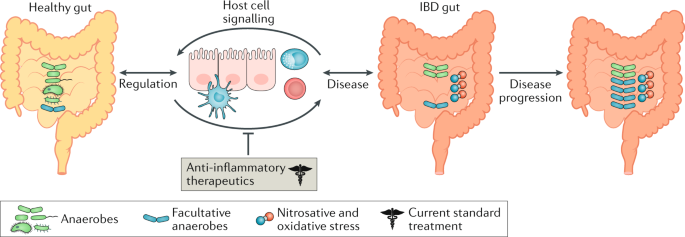
Microbial genes and pathways in inflammatory bowel disease
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Perturbations in the intestinal microbiome are implicated in inflammatory bowel disease (IBD). Studies of treatment-naive patients have identified microbial taxa associated with
disease course and treatment efficacy. To gain a mechanistic understanding of how the microbiome affects gastrointestinal health, we need to move from census to function. Bacteria, including
those that adhere to epithelial cells as well as several _Clostridium_ species, can alter differentiation of T helper 17 cells and regulatory T cells. Similarly, microbial products such as
short-chain fatty acids and sphingolipids also influence immune responses. Metagenomics and culturomics have identified strains of _Ruminococcus gnavus_ and adherent invasive _Escherichia
coli_ that are linked to IBD and gut inflammation. Integrated analysis of multiomics data, including metagenomics, metatranscriptomics and metabolomics, with measurements of host response
and culturomics, have great potential in understanding the role of the microbiome in IBD. In this Review, we highlight current knowledge of gut microbial factors linked to IBD pathogenesis
and discuss how multiomics data from large-scale population studies in health and disease have been used to identify specific microbial strains, transcriptional changes and metabolic
alterations associated with IBD. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal
Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices
may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support
SIMILAR CONTENT BEING VIEWED BY OTHERS EXPLORING HOW MICROBIOME SIGNATURES CHANGE ACROSS INFLAMMATORY BOWEL DISEASE CONDITIONS AND DISEASE LOCATIONS Article Open access 21 September 2021
MICROBIOTA IN INFLAMMATORY BOWEL DISEASE: MECHANISMS OF DISEASE AND THERAPEUTIC OPPORTUNITIES Article 10 March 2025 ASSOCIATION OF DISTINCT MICROBIAL AND METABOLIC SIGNATURES WITH
MICROSCOPIC COLITIS Article Open access 23 May 2025 REFERENCES * Ijssennagger, N. et al. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus
barrier in colon. _Proc. Natl Acad. Sci. USA_ 112, 10038–10043 (2015). Article CAS PubMed PubMed Central Google Scholar * Yano, J. M. et al. Indigenous bacteria from the gut microbiota
regulate host serotonin biosynthesis. _Cell_ 161, 264–276 (2015). Article CAS PubMed PubMed Central Google Scholar * Reinhardt, C. et al. Tissue factor and PAR1 promote
microbiota-induced intestinal vascular remodelling. _Nature_ 483, 627–631 (2012). Article CAS PubMed Google Scholar * Cho, I. et al. Antibiotics in early life alter the murine colonic
microbiome and adiposity. _Nature_ 488, 621–626 (2012). Article CAS PubMed PubMed Central Google Scholar * Lee, W. J. & Hase, K. Gut microbiota-generated metabolites in animal
health and disease. _Nat. Chem. Biol._ 10, 416–424 (2014). Article CAS PubMed Google Scholar * Fischbach, M. A. & Segre, J. A. Signaling in host-associated microbial communities.
_Cell_ 164, 1288–1300 (2016). Article CAS PubMed PubMed Central Google Scholar * Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. _Nature_ 457, 480–484 (2009).
Article CAS PubMed Google Scholar * Le Chatelier, E. et al. Richness of human gut microbiome correlates with metabolic markers. _Nature_ 500, 541–546 (2013). Article PubMed CAS Google
Scholar * Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. _Nature_ 490, 55–60 (2012). Article CAS PubMed Google Scholar * Karlsson, F. H. et
al. Gut metagenome in European women with normal, impaired and diabetic glucose control. _Nature_ 498, 99–103 (2013). Article CAS PubMed Google Scholar * Wang, Z. et al. Gut flora
metabolism of phosphatidylcholine promotes cardiovascular disease. _Nature_ 472, 57–63 (2011). Article CAS PubMed PubMed Central Google Scholar * Koeth, R. A. et al. Intestinal
microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. _Nat. Med._ 19, 576–585 (2013). Article CAS PubMed PubMed Central Google Scholar * Tang, W. H. et
al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. _N. Engl. J. Med._ 368, 1575–1584 (2013). Article CAS PubMed PubMed Central Google Scholar * Hsiao,
E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. _Cell_ 155, 1451–1463 (2013). Article CAS PubMed PubMed Central
Google Scholar * Fujimura, K. E. et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. _Nat. Med._ 22, 1187–1191 (2016). Article CAS
PubMed PubMed Central Google Scholar * Arrieta, M. C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. _Sci. Transl Med._ 7, 307ra152 (2015).
Article PubMed CAS Google Scholar * Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. _Cell_ 165, 842–853 (2016). Article CAS PubMed
PubMed Central Google Scholar * Zhao, G. et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. _Proc. Natl Acad. Sci. USA_ 114, E6166–E6175
(2017). CAS PubMed PubMed Central Google Scholar * Huang, H. et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. _Nature_ 547, 173–178 (2017). Article CAS
PubMed PubMed Central Google Scholar * Lamas, B. et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. _Nat. Med._ 22,
598–605 (2016). Article CAS PubMed PubMed Central Google Scholar * Imhann, F. et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of
inflammatory bowel disease. _Gut_ 67, 108–119 (2018). Article CAS PubMed Google Scholar * Ng, S. C. et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st
century: a systematic review of population-based studies. _Lancet_ 390, 2769–2778 (2018). Article Google Scholar * Zhernakova, A. et al. Population-based metagenomics analysis reveals
markers for gut microbiome composition and diversity. _Science_ 352, 565–569 (2016). Article CAS PubMed PubMed Central Google Scholar * Rothschild, D. et al. Environment dominates over
host genetics in shaping human gut microbiota. _Nature_ 555, 210–215 (2018). Article CAS PubMed Google Scholar * Kronman, M. P., Zaoutis, T. E., Haynes, K., Feng, R. & Coffin, S. E.
Antibiotic exposure and IBD development among children: a population-based cohort study. _Pediatrics_ 130, e794–e803 (2012). Article PubMed PubMed Central Google Scholar * Ferrante, M.
et al. New serological markers in inflammatory bowel disease are associated with complicated disease behaviour. _Gut_ 56, 1394–1403 (2007). Article PubMed PubMed Central Google Scholar *
Dotan, I. et al. Antibodies against laminaribioside and chitobioside are novel serologic markers in Crohn’s disease. _Gastroenterology_ 131, 366–378 (2006). Article CAS PubMed Google
Scholar * Schirmer, M. et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. _Cell Host Microbe_ 24, 600–610
(2018). Article CAS PubMed PubMed Central Google Scholar * Schaubeck, M. et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in
antimicrobial defence. _Gut_ 65, 225–237 (2016). Article CAS PubMed Google Scholar * Couturier-Maillard, A. et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and
colorectal cancer. _J. Clin. Invest._ 123, 700–711 (2013). CAS PubMed PubMed Central Google Scholar * Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk
for colitis. _Cell_ 145, 745–757 (2011). Article CAS PubMed PubMed Central Google Scholar * Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the
innate immune system. _Cell_ 131, 33–45 (2007). Article CAS PubMed PubMed Central Google Scholar * Henao-Mejia, J. et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD
and obesity. _Nature_ 482, 179–185 (2012). Article CAS PubMed PubMed Central Google Scholar * Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria.
_Cell_ 139, 485–498 (2009). Article CAS PubMed PubMed Central Google Scholar * Sellon, R. K. et al. Resident enteric bacteria are necessary for development of spontaneous colitis and
immune system activation in interleukin-10-deficient mice. _Infect. Immun._ 66, 5224–5231 (1998). Article CAS PubMed PubMed Central Google Scholar * Schultz, M. et al. IL-2-deficient
mice raised under germfree conditions develop delayed mild focal intestinal inflammation. _Am. J. Physiol._ 276, G1461–G1472 (1999). CAS PubMed Google Scholar * Rath, H. C., Wilson, K. H.
& Sartor, R. B. Differential induction of colitis and gastritis in HLA-B27 transgenic rats selectively colonized with _Bacteroides vulgatus_ or _Escherichia coli_. _Infect. Immun._ 67,
2969–2974 (1999). Article CAS PubMed PubMed Central Google Scholar * Xavier, R. J. & Podolsky, D. K. Unravelling the pathogenesis of inflammatory bowel disease. _Nature_ 448,
427–434 (2007). Article CAS PubMed Google Scholar * Bunker, J. J. & Bendelac, A. IgA responses to microbiota. _Immunity_ 49, 211–224 (2018). Article CAS PubMed PubMed Central
Google Scholar * Mkaddem, S. B. et al. IgA, IgA receptors, and their anti-inflammatory properties. _Curr. Top. Microbiol. Immunol._ 382, 221–235 (2014). PubMed Google Scholar * Moon, C.
et al. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. _Nature_ 521, 90–93 (2015). Article CAS PubMed PubMed Central Google Scholar * Mohanan,
V. et al. C1orf106 is a colitis risk gene that regulates stability of epithelial adherens junctions. _Science_ 359, 1161–1166 (2018). Article CAS PubMed PubMed Central Google Scholar *
Gaudier, E. et al. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. _Am. J. Physiol. Gastrointest. Liver Physiol._ 287,
G1168–G1174 (2004). Article CAS PubMed Google Scholar * Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. _Nature_ 504,
446–450 (2013). Article CAS PubMed Google Scholar * Kim, M. H., Kang, S. G., Park, J. H., Yanagisawa, M. & Kim, C. H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal
epithelial cells to promote inflammatory responses in mice. _Gastroenterology_ 145, 396–406 (2013). Article CAS PubMed Google Scholar * Venkatesh, M. et al. Symbiotic bacterial
metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. _Immunity_ 41, 296–310 (2014). Article CAS PubMed PubMed Central Google
Scholar * Wlodarska, M. et al. Indoleacrylic acid produced by commensal _Peptostreptococcus_ species suppresses inflammation. _Cell Host Microbe_ 22, 25–37 (2017). THIS WORK IDENTIFIES A
TRYPTOPHAN DERIVATIVE PRODUCED BY GUT BACTERIA THAT INCREASES MUCUS PRODUCTION AND DECREASES INFLAMMATORY CYTOKINE PRODUCTION. Article CAS PubMed PubMed Central Google Scholar *
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. _Proc. Natl Acad. Sci. USA_ 107,
11971–11975 (2010). Article PubMed PubMed Central Google Scholar * Backhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. _Cell Host
Microbe_ 17, 690–703 (2015). Article PubMed CAS Google Scholar * Jakobsson, H. E. et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses
in infants delivered by caesarean section. _Gut_ 63, 559–566 (2014). Article CAS PubMed Google Scholar * Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous
_Clostridium_ species. _Science_ 331, 337–341 (2011). Article CAS PubMed Google Scholar * Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer
T cell function. _Science_ 336, 489–493 (2012). Article CAS PubMed PubMed Central Google Scholar * An, D. et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host
intestinal natural killer T cells. _Cell_ 156, 123–133 (2014). THIS PAPER SHOWS THAT SPHINGOLIPIDS PRODUCED BY INTESTINAL BACTERIA HAVE A ROLE IN COLONIC INKT CELL HOMEOSTASIS. Article CAS
PubMed PubMed Central Google Scholar * Lopez-Serrano, P. et al. Environmental risk factors in inflammatory bowel diseases. Investigating the hygiene hypothesis: a Spanish case–control
study. _Scand. J. Gastroenterol._ 45, 1464–1471 (2010). Article PubMed Google Scholar * von Mutius, E. Allergies, infections and the hygiene hypothesis—the epidemiological evidence.
_Immunobiology_ 212, 433–439 (2007). Article CAS Google Scholar * Britton, G. J. et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORγt+
regulatory T cells and exacerbate colitis in mice. _Immunity_ 50, 212–224 (2019). Article CAS PubMed PubMed Central Google Scholar * Gaboriau-Routhiau, V. et al. The key role of
segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. _Immunity_ 31, 677–689 (2009). Article CAS PubMed Google Scholar * Mao, K. et al. Innate and
adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. _Nature_ 554, 255–259 (2018). Article CAS PubMed Google Scholar * Atarashi, K. et al. Th17 cell induction
by adhesion of microbes to intestinal epithelial cells. _Cell_ 163, 367–380 (2015). Article CAS PubMed PubMed Central Google Scholar * Dobes, J. et al. Gastrointestinal autoimmunity
associated with loss of central tolerance to enteric α-defensins. _Gastroenterology_ 149, 139–150 (2015). Article CAS PubMed Google Scholar * Round, J. L. & Mazmanian, S. K.
Inducible Foxp3+ regulatory T cell development by a commensal bacterium of the intestinal microbiota. _Proc. Natl Acad. Sci. USA_ 107, 12204–12209 (2010). Article CAS PubMed PubMed
Central Google Scholar * Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. _Nature_ 500, 232–236 (2013). Article CAS
PubMed Google Scholar * Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn’s disease. _Cell Host Microbe_ 15, 382–392 (2014). Article CAS PubMed PubMed Central Google
Scholar * Sokol, H., Lay, C., Seksik, P. & Tannock, G. W. Analysis of bacterial bowel communities of IBD patients: what has it revealed? _Inflamm. Bowel Dis._ 14, 858–867 (2008).
Article PubMed Google Scholar * Scanlan, P. D., Shanahan, F., O’Mahony, C. & Marchesi, J. R. Culture-independent analyses of temporal variation of the dominant fecal microbiota and
targeted bacterial subgroups in Crohn’s disease. _J. Clin. Microbiol._ 44, 3980–3988 (2006). Article CAS PubMed PubMed Central Google Scholar * Ott, S. J. et al. Reduction in diversity
of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. _Gut_ 53, 685–693 (2004). Article CAS PubMed PubMed Central Google Scholar *
Gophna, U., Sommerfeld, K., Gophna, S., Doolittle, W. F. & Veldhuyzen van Zanten, S. J. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and
ulcerative colitis. _J. Clin. Microbiol._ 44, 4136–4141 (2006). Article CAS PubMed PubMed Central Google Scholar * Frank, D. N. et al. Molecular-phylogenetic characterization of
microbial community imbalances in human inflammatory bowel diseases. _Proc. Natl Acad. Sci. USA_ 104, 13780–13785 (2007). Article CAS PubMed PubMed Central Google Scholar * Swidsinski,
A., Weber, J., Loening-Baucke, V., Hale, L. P. & Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. _J. Clin. Microbiol._
43, 3380–3389 (2005). Article PubMed PubMed Central Google Scholar * Swidsinski, A. et al. Mucosal flora in inflammatory bowel disease. _Gastroenterology_ 122, 44–54 (2002). Article
PubMed Google Scholar * Kotlowski, R., Bernstein, C. N., Sepehri, S. & Krause, D. O. High prevalence of _Escherichia coli_ belonging to the B2+D phylogenetic group in inflammatory
bowel disease. _Gut_ 56, 669–675 (2007). Article CAS PubMed Google Scholar * Mylonaki, M., Rayment, N. B., Rampton, D. S., Hudspith, B. N. & Brostoff, J. Molecular characterization
of rectal mucosa-associated bacterial flora in inflammatory bowel disease. _Inflamm. Bowel Dis._ 11, 481–487 (2005). Article PubMed Google Scholar * Conte, M. P. et al. Gut-associated
bacterial microbiota in paediatric patients with inflammatory bowel disease. _Gut_ 55, 1760–1767 (2006). Article CAS PubMed PubMed Central Google Scholar * Schultsz, C., Van Den Berg,
F. M., Ten Kate, F. W., Tytgat, G. N. & Dankert, J. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls.
_Gastroenterology_ 117, 1089–1097 (1999). Article CAS PubMed Google Scholar * Prindiville, T., Cantrell, M. & Wilson, K. H. Ribosomal DNA sequence analysis of mucosa-associated
bacteria in Crohn’s disease. _Inflamm. Bowel Dis._ 10, 824–833 (2004). Article PubMed Google Scholar * Yilmaz, B. et al. Microbial network disturbances in relapsing refractory Crohn’s
disease. _Nat. Med._ 25, 323–336 (2019). Article CAS PubMed Google Scholar * Morgan, X. C. et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment.
_Genome Biol._ 13, R79 (2012). Article CAS PubMed PubMed Central Google Scholar * Vich Vila, A. et al. Gut microbiota composition and functional changes in inflammatory bowel disease
and irritable bowel syndrome. _Sci. Transl Med._ 10, eaap8914 (2018). Article PubMed CAS Google Scholar * Franzosa, E. A. et al. Gut microbiome structure and metabolic activity in
inflammatory bowel disease. _Nat. Microbiol._ 4, 293–305 (2019). THIS WORK PAIRS METAGENOMIC AND METABOLOMIC DATA FROM A CROSS-SECTIONAL IBD COHORT TO IDENTIFY ASSOCIATIONS BETWEEN GUT
BACTERIA AND METABOLITES THAT ARE PREDICTIVE OF IBD STATUS. Article CAS PubMed Google Scholar * Lewis, J. D. et al. Inflammation, antibiotics, and diet as environmental stressors of the
gut microbiome in pediatric Crohn’s disease. _Cell Host Microbe_ 18, 489–500 (2015). THIS STUDY SHOWS THAT MICROBIAL DYSBIOSIS RESULTS FROM INDEPENDENT EFFECTS OF INFLAMMATION, DIET AND
ANTIBIOTICS AND IS AN IMPORTANT FACTOR IN THE CONTEXT OF ENTERAL NUTRITION AND ANTI-TNF TREATMENT IN PAEDIATRIC PATIENTS WITH CD. Article CAS PubMed PubMed Central Google Scholar *
Hall, A. B. et al. A novel _Ruminococcus gnavus_ clade enriched in inflammatory bowel disease patients. _Genome Med._ 9, 103 (2017). Article PubMed PubMed Central CAS Google Scholar *
The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. _Nature_ 486, 207–214 (2012). Article PubMed Central CAS Google Scholar *
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. _Nature_ 569, 655–662 (2019) * Sundin, J. et al. Fecal chromogranins and secretogranins are
linked to the fecal and mucosal intestinal bacterial composition of IBS patients and healthy subjects. _Sci. Rep._ 8, 16821 (2018). Article PubMed PubMed Central CAS Google Scholar *
Konstantinidis, K. T., Rossello-Mora, R. & Amann, R. Uncultivated microbes in need of their own taxonomy. _ISME J._ 11, 2399–2406 (2017). Article PubMed PubMed Central Google Scholar
* Martinez-Medina, M. & Garcia-Gil, L. J. _Escherichia coli_ in chronic inflammatory bowel diseases: an update on adherent invasive _Escherichia coli_ pathogenicity. _World J.
Gastrointest. Pathophysiol._ 5, 213–227 (2014). Article PubMed PubMed Central Google Scholar * Elhenawy, W., Tsai, C. N. & Coombes, B. K. Host-specific adaptive diversification of
Crohn’s disease-associated adherent-invasive _Escherichia coli_. _Cell Host Microbe_ 25, 301–312 (2019). Article CAS PubMed Google Scholar * Palmela, C. et al. Adherent-invasive
_Escherichia coli_ in inflammatory bowel disease. _Gut_ 67, 574–587 (2018). Article CAS PubMed Google Scholar * Atarashi, K. et al. Ectopic colonization of oral bacteria in the intestine
drives TH1 cell induction and inflammation. _Science_ 358, 359–365 (2017). THIS STUDY LINKS SPECIFIC STRAINS OF ORAL _KLEBSIELLA_ SPECIES THAT ARE INCREASED IN IBD WITH T H 1 CELL INDUCTION
AND INFLAMMATION. Article CAS PubMed PubMed Central Google Scholar * Rashid, T., Ebringer, A. & Wilson, C. The role of _Klebsiella_ in Crohn’s disease with a potential for the use
of antimicrobial measures. _Int. J. Rheumatol._ 2013, 610393 (2013). Article PubMed PubMed Central Google Scholar * Garrett, W. S. et al. Enterobacteriaceae act in concert with the gut
microbiota to induce spontaneous and maternally transmitted colitis. _Cell Host Microbe_ 8, 292–300 (2010). Article CAS PubMed PubMed Central Google Scholar * Carvalho, F. A. et al.
Crohn’s disease adherent-invasive _Escherichia coli_ colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. _J. Exp. Med._ 206, 2179–2189 (2009). Article
CAS PubMed PubMed Central Google Scholar * Lopez-Siles, M., Duncan, S. H., Garcia-Gil, L. J. & Martinez-Medina, M. _Faecalibacterium prausnitzii_: from microbiology to diagnostics
and prognostics. _ISME J._ 11, 841–852 (2017). Article PubMed PubMed Central Google Scholar * Franzosa, E. A. et al. Relating the metatranscriptome and metagenome of the human gut.
_Proc. Natl Acad. Sci. USA_ 111, E2329–E2338 (2014). Article CAS PubMed PubMed Central Google Scholar * Schirmer, M. et al. Dynamics of metatranscription in the inflammatory bowel
disease gut microbiome. _Nat. Microbiol._ 3, 337–346 (2018). THIS WORK DEMONSTRATES THAT METATRANSCRIPTOMIC PROFILING CAN REVEAL SPECIES-SPECIFIC BIASES IN TRANSCRIPTIONAL ACTIVITY OF GUT
BACTERIA, INCLUDING IBD-SPECIFIC MICROBIAL CHARACTERISTICS, PROVIDING NEW INSIGHT INTO THE POTENTIAL MECHANISMS OF HOST–MICROBIAL DYSBIOSIS IN DISEASE. Article CAS PubMed PubMed Central
Google Scholar * Korem, T. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. _Science_ 349, 1101–1106 (2015). Article CAS PubMed
PubMed Central Google Scholar * Thorburn, A. N., Macia, L. & Mackay, C. R. Diet, metabolites, and “western-lifestyle” inflammatory diseases. _Immunity_ 40, 833–842 (2014). Article CAS
PubMed Google Scholar * Donia, M. S. & Fischbach, M. A. Human microbiota. Small molecules from the human microbiota. _Science_ 349, 1254766 (2015). Article PubMed PubMed Central
CAS Google Scholar * Ursell, L. K. et al. The intestinal metabolome: an intersection between microbiota and host. _Gastroenterology_ 146, 1470–1476 (2014). Article CAS PubMed Google
Scholar * Levy, M. et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. _Cell_ 163, 1428–1443 (2015). Article CAS
PubMed PubMed Central Google Scholar * Jacobs, J. P. et al. A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. _Cell.
Mol. Gastroenterol. Hepatol._ 2, 750–766 (2016). Article PubMed PubMed Central Google Scholar * Kolho, K. L., Pessia, A., Jaakkola, T., de Vos, W. M. & Velagapudi, V. Faecal and
serum metabolomics in paediatric inflammatory bowel disease. _J. Crohns Colitis_ 11, 321–334 (2017). Article PubMed Google Scholar * Scoville, E. A. et al. Alterations in lipid, amino
acid, and energy metabolism distinguish Crohn’s disease from ulcerative colitis and control subjects by serum metabolomic profiling. _Metabolomics_ 14, 17 (2018). Article PubMed CAS
Google Scholar * Jansson, J. et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. _PLOS ONE_ 4, e6386 (2009). Article PubMed PubMed Central CAS Google Scholar *
Mitsuyama, K. et al. Antibody markers in the diagnosis of inflammatory bowel disease. _World J. Gastroenterol._ 22, 1304–1310 (2016). Article CAS PubMed PubMed Central Google Scholar *
Sokol, H. et al. Fungal microbiota dysbiosis in IBD. _Gut_ 66, 1039–1048 (2017). Article CAS PubMed Google Scholar * Leonardi, I. et al. CX3CR1+ mononuclear phagocytes control immunity
to intestinal fungi. _Science_ 359, 232–236 (2018). Article CAS PubMed PubMed Central Google Scholar * Chiaro, T. R. et al. A member of the gut mycobiota modulates host purine
metabolism exacerbating colitis in mice. _Sci. Transl Med._ 9, eaaf9044 (2017). Article PubMed PubMed Central CAS Google Scholar * Limon, J. J. et al. Malassezia is associated with
Crohn’s disease and exacerbates colitis in mouse models. _Cell Host Microbe_ 25, 377–388 (2019). Article CAS PubMed PubMed Central Google Scholar * Cadwell, K. et al.
Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. _Cell_ 141, 1135–1145 (2010). Article CAS PubMed PubMed Central Google Scholar
* McGovern, D. P. et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. _Hum. Mol. Genet._ 19, 3468–3476 (2010). Article CAS PubMed PubMed Central
Google Scholar * Wilen, C. B. et al. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. _Science_ 360, 204–208 (2018). Article CAS PubMed PubMed Central
Google Scholar * Norman, J. M. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. _Cell_ 160, 447–460 (2015). Article CAS PubMed PubMed Central
Google Scholar * Lopetuso, L. R., Ianiro, G., Scaldaferri, F., Cammarota, G. & Gasbarrini, A. Gut virome and inflammatory bowel disease. _Inflamm. Bowel Dis._ 22, 1708–1712 (2016).
Article PubMed Google Scholar * Ananthakrishnan, A. N. et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. _Cell Host
Microbe_ 21, 603–610 (2017). Article CAS PubMed PubMed Central Google Scholar * Kolho, K. L. et al. Fecal microbiota in pediatric inflammatory bowel disease and its relation to
inflammation. _Am. J. Gastroenterol._ 110, 921–930 (2015). Article PubMed Google Scholar * Doherty, M. K. et al. Fecal microbiota signatures are associated with response to ustekinumab
therapy among Crohn’s disease patients. _mBio_ 9, e02120–17 (2018). Article CAS PubMed PubMed Central Google Scholar * Shaw, K. A. et al. Dysbiosis, inflammation, and response to
treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. _Genome Med._ 8, 75 (2016). Article PubMed PubMed Central CAS Google Scholar *
Halmos, E. P. & Gibson, P. R. Dietary management of IBD—insights and advice. _Nat. Rev. Gastroenterol. Hepatol._ 12, 133–146 (2015). Article CAS PubMed Google Scholar * Narula, N. et
al. Enteral nutritional therapy for induction of remission in Crohn’s disease. _Cochrane Database Syst. Rev._ 4, CD000542 (2018). PubMed Google Scholar * Dziechciarz, P., Horvath, A.,
Shamir, R. & Szajewska, H. Meta-analysis: enteral nutrition in active Crohn’s disease in children. _Aliment. Pharmacol. Ther._ 26, 795–806 (2007). Article CAS PubMed Google Scholar *
Albenberg, L. G. & Wu, G. D. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. _Gastroenterology_ 146, 1564–1572 (2014). Article CAS
PubMed Google Scholar * Lee, D. et al. Diet in the pathogenesis and treatment of inflammatory bowel diseases. _Gastroenterology_ 148, 1087–1106 (2015). Article CAS PubMed Google
Scholar * Ananthakrishnan, A. N. et al. A prospective study of long-term intake of dietary fiber and risk of Crohn’s disease and ulcerative colitis. _Gastroenterology_ 145, 970–977 (2013).
Article CAS PubMed Google Scholar * Llewellyn, S. R. et al. Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice.
_Gastroenterology_ 154, 1037–1046 (2018). Article PubMed Google Scholar * Albenberg, L. et al. A diet low in red and processed meat does not reduce rate of Crohn’s disease flares.
_Gastroenterology_ https://doi.org/10.1053/j.gastro.2019.03.015 (2019). Article PubMed Google Scholar * Paramsothy, S. et al. Faecal microbiota transplantation for inflammatory bowel
disease: a systematic review and meta-analysis. _J. Crohns Colitis_ 11, 1180–1199 (2017). Article PubMed Google Scholar * Levy, A. N. & Allegretti, J. R. Insights into the role of
fecal microbiota transplantation for the treatment of inflammatory bowel disease. _Therap. Adv. Gastroenterol._ 12, 1756284819836893 (2019). Article PubMed PubMed Central Google Scholar
* Smillie, C. S. et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. _Cell Host Microbe_ 23, 229–240 (2018).
Article CAS PubMed PubMed Central Google Scholar * Vermeire, S. et al. Donor species richness determines faecal microbiota transplantation success in inflammatory bowel disease. _J.
Crohns Colitis_ 10, 387–394 (2016). Article PubMed Google Scholar * Palm, N. W. et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. _Cell_ 158,
1000–1010 (2014). THIS STUDY PIONEERED THE USE OF IGA COATING TO IDENTIFY INFLAMMATORY MEMBERS OF THE IBD GUT MICROBIOTA. Article CAS PubMed PubMed Central Google Scholar * Larmonier,
C. B., Shehab, K. W., Ghishan, F. K. & Kiela, P. R. T. Lymphocyte dynamics in inflammatory bowel diseases: role of the microbiome. _Biomed. Res. Int._ 2015, 504638 (2015). Article CAS
PubMed PubMed Central Google Scholar * Ni, J. et al. A role for bacterial urease in gut dysbiosis and Crohn’s disease. _Sci. Transl Med._ 9, eaah6888 (2017). Article PubMed PubMed
Central CAS Google Scholar * Winter, S. E. et al. Host-derived nitrate boosts growth of _E. coli_ in the inflamed gut. _Science_ 339, 708–711 (2013). Article CAS PubMed PubMed Central
Google Scholar * Zhu, W. et al. Precision editing of the gut microbiota ameliorates colitis. _Nature_ 553, 208–211 (2018). THIS PAPER SHOWS THAT TUNGSTATE TREATMENT INHIBITS
MOLYBDENUM-COFACTOR-DEPENDENT MICROBIAL RESPIRATORY PATHWAYS IN THE MAMMALIAN GUT AND PREVENTS THE DYSBIOTIC EXPANSION OF ENTEROBACTERIACEAE DURING GUT INFLAMMATION. Article CAS PubMed
PubMed Central Google Scholar * Kugathasan, S. et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study.
_Lancet_ 389, 1710–1718 (2017). Article PubMed PubMed Central Google Scholar * Hyams, J. S. et al. Clinical and biological predictors of response to standardised paediatric colitis
therapy (PROTECT): a multicentre inception cohort study. _Lancet_ 393, 1708–1720 (2019). Article PubMed PubMed Central Google Scholar * Tanoue, T. et al. A defined commensal consortium
elicits CD8 T cells and anti-cancer immunity. _Nature_ 565, 600–605 (2019). Article CAS PubMed Google Scholar * Podolsky, D. K. Inflammatory bowel disease. _N. Engl. J. Med._ 347,
417–429 (2002). Article CAS PubMed Google Scholar * Hering, N. A., Fromm, M. & Schulzke, J. D. Determinants of colonic barrier function in inflammatory bowel disease and potential
therapeutics. _J. Physiol._ 590, 1035–1044 (2012). Article CAS PubMed PubMed Central Google Scholar * Salzman, N. H. et al. Enteric defensins are essential regulators of intestinal
microbial ecology. _Nat. Immunol._ 11, 76–83 (2010). Article CAS PubMed Google Scholar * VanDussen, K. L. et al. Genetic variants synthesize to produce Paneth cell phenotypes that define
subtypes of Crohn’s disease. _Gastroenterology_ 146, 200–209 (2014). Article CAS PubMed Google Scholar * Adolph, T. E. et al. Paneth cells as a site of origin for intestinal
inflammation. _Nature_ 503, 272–276 (2013). Article CAS PubMed PubMed Central Google Scholar * Surawicz, C. M., Haggitt, R. C., Husseman, M. & McFarland, L. V. Mucosal biopsy
diagnosis of colitis: acute self-limited colitis and idiopathic inflammatory bowel disease. _Gastroenterology_ 107, 755–763 (1994). Article CAS PubMed Google Scholar * Van der Sluis, M.
et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. _Gastroenterology_ 131, 117–129 (2006). Article CAS PubMed Google
Scholar * McDole, J. R. et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. _Nature_ 483, 345–349 (2012). Article CAS PubMed PubMed Central
Google Scholar * Desai, D., Faubion, W. A. & Sandborn, W. J. Review article: biological activity markers in inflammatory bowel disease. _Aliment. Pharmacol. Ther._ 25, 247–255 (2007).
Article CAS PubMed Google Scholar * Francescone, R., Hou, V. & Grivennikov, S. I. Cytokines, IBD, and colitis-associated cancer. _Inflamm. Bowel Dis._ 21, 409–418 (2015). Article
PubMed Google Scholar * Fernando, M. R., Saxena, A., Reyes, J. L. & McKay, D. M. Butyrate enhances antibacterial effects while suppressing other features of alternative activation in
IL-4-induced macrophages. _Am. J. Physiol. Gastrointest. Liver Physiol._ 310, G822–G831 (2016). Article PubMed Google Scholar * Wikoff, W. R. et al. Metabolomics analysis reveals large
effects of gut microflora on mammalian blood metabolites. _Proc. Natl Acad. Sci. USA_ 106, 3698–3703 (2009). Article CAS PubMed PubMed Central Google Scholar * Zelante, T. et al.
Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. _Immunity_ 39, 372–385 (2013). Article CAS PubMed Google Scholar
* Williams, B. B. et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. _Cell Host Microbe_ 16, 495–503 (2014). Article
CAS PubMed PubMed Central Google Scholar * Devlin, A. S. & Fischbach, M. A. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. _Nat. Chem. Biol._ 11,
685–690 (2015). Article CAS PubMed PubMed Central Google Scholar * Cao, W. et al. The xenobiotic transporter Mdr1 enforces T cell homeostasis in the presence of intestinal bile acids.
_Immunity_ 47, 1182–1196 (2017). Article CAS PubMed PubMed Central Google Scholar * Fischbach, M. A. & Sonnenburg, J. L. Eating for two: how metabolism establishes interspecies
interactions in the gut. _Cell Host Microbe_ 10, 336–347 (2011). Article CAS PubMed PubMed Central Google Scholar * Tannahill, G. M. et al. Succinate is an inflammatory signal that
induces IL-1β through HIF-1α. _Nature_ 496, 238–242 (2013). Article CAS PubMed PubMed Central Google Scholar * Serena, C. et al. Elevated circulating levels of succinate in human
obesity are linked to specific gut microbiota. _ISME J._ 12, 1642–1657 (2018). Article CAS PubMed PubMed Central Google Scholar * Ferreyra, J. A. et al. Gut microbiota-produced
succinate promotes _C. difficile_ infection after antibiotic treatment or motility disturbance. _Cell Host Microbe_ 16, 770–777 (2014). Article CAS PubMed PubMed Central Google Scholar
* De Vadder, F. et al. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. _Cell Metab._ 24, 151–157 (2016). Article PubMed CAS Google Scholar *
Abdel Hadi, L., Di Vito, C. & Riboni, L. Fostering inflammatory bowel disease: sphingolipid strategies to join forces. _Mediators Inflamm._ 2016, 3827684 (2016). Article PubMed PubMed
Central CAS Google Scholar * Segata, N. GraPhlAn. _Bitbucket_ https://bitbucket.org/nsegata/graphlan/wiki/Home (2018). * Vaughn, B. P. et al. Increased intestinal microbial diversity
following fecal microbiota transplant for active Crohn’s disease. _Inflamm. Bowel Dis._ 22, 2182–2190 (2016). Article PubMed Google Scholar Download references ACKNOWLEDGEMENTS The
authors thank T. Reimels for editorial assistance and for help with figure design. R.J.X. received funding from the US National Institutes of Health (P30 DK043351 and R01 AT009708), the
Crohn’s and Colitis Foundation of America, and the Center for Microbiome Informatics and Therapeutics at MIT. REVIEWER INFORMATION _Nature Reviews Microbiology_ thanks J. Faith, C.
Manichanh, and other anonymous reviewer(s), for their contribution to the peer review of this work. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Broad Institute of MIT and Harvard,
Cambridge, MA, USA Melanie Schirmer, Ashley Garner, Hera Vlamakis & Ramnik J. Xavier * Center for Microbiome Informatics and Therapeutics, MIT, Cambridge, MA, USA Hera Vlamakis &
Ramnik J. Xavier Authors * Melanie Schirmer View author publications You can also search for this author inPubMed Google Scholar * Ashley Garner View author publications You can also search
for this author inPubMed Google Scholar * Hera Vlamakis View author publications You can also search for this author inPubMed Google Scholar * Ramnik J. Xavier View author publications You
can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.S., A.G., H.V., and R.J.X. conceived and wrote the article. All authors substantially contributed to discussion of
content and reviewed/edited the manuscript before submission. M.S. generated Fig. 2. CORRESPONDING AUTHORS Correspondence to Melanie Schirmer, Hera Vlamakis or Ramnik J. Xavier. ETHICS
DECLARATIONS COMPETING INTERESTS R.J.X. is a consultant to Nestle and Novartis. All other authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature
remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RELATED LINKS INFLAMMATORY BOWEL DISEASE MULTI’OMICS DATABASE. https://ibdmdb.org/
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION GLOSSARY * Microbiota The collection of microorganisms in a particular environment. * Microbiome The genes, genomes and products of the
microbiota. * Barrier function Epithelial cell–cell junctions plus the mucosal layer that permit nutrients and prevent luminal contents from accessing the rest of the body. * Glycaemic
response The glycaemic response to food describes its effect on blood glucose levels after consumption. * Non-caseating granulomas Granulomas are clusters of immune cells that form during
infection, inflammation or in the presence of a foreign substance to prevent a systemic spread. The absence of necrosis is a defining feature of non-caseating granulomas. In Crohn’s disease,
non-caseating granulomas are formed during inflammation without an obvious infectious trigger. * Hygiene hypothesis According to the hygiene hypothesis, a lack of early childhood microbial
exposure affects the development of the immune system. This has been ascribed to the increase of allergic and autoimmune diseases in Western countries. * Mannan A mannose polymer and
component of fungal and plant cell walls. * Atypical perinuclear anti-neutrophil cytoplasmic antibody Anti-neutrophil cytoplasmic antibodies are classified based on staining patterns.
Cytoplasmic anti-neutrophil cytoplasmic antibody (ANCA) refers to staining of the entire cytoplasm, and perinuclear ANCA refers to staining of the area around the nucleus. Perinuclear ANCAs
have been implicated in inflammatory bowel disease; however, their target antigens are unknown and they are therefore described as atypical perinuclear ANCA. * Laminaribioside A glucose
disaccharide building block of laminarin and a component of the cell walls of fungi and algae. * Chitobioside A building block of the _N_-acetylglucosamine-based glycan chitin and a
component of the cell walls of microorganisms. * Mannobioside A disaccharide of mannose. * Colectomy A surgical procedure removing all or part of the colon. * Strains The classical
microbiological definition of strain is a single bacterial isolate. In the context of metagenomic data, it refers to a combination of single-nucleotide polymorphisms that are computationally
predicted to be linked and originating from an individual strain genome. * Metatranscriptome All of the RNA in an environment. * Culturomics The process of using classical microbiological
techniques to culture and identify unknown bacteria that inhabit a given environment. * Indole metabolites Indole metabolites derive from microbial metabolism of tryptophan and can be
recognized by several host receptors that regulate host–microbial homeostasis. * Metagenomes All the genetic material present in an environment, consisting of the genomes of numerous
organisms. * RORγt+ Treg cells Regulatory T cells that express the transcription factor RORγt, a nuclear hormone receptor and critical regulator of antimicrobial immunity. * Autoimmune
polyendocrinopathy–candidiasis–ectodermal dystrophy An autoimmune disease characterized by destruction of endocrine tissues, chronic mucocutaneous candidiasis and ectodermal disorders. * 16S
ribosomal RNA (rRNA) gene A gene conserved among bacteria often used for taxonomic classification. * Keystone taxa Species with high connectivity in microbial networks (built based on
statistical associations), suggesting that they are a key component of the ecosystem and their removal would result in drastic changes to the microbial ecosystem. * Anti-tumour necrosis
factor therapy Drugs that target tumour necrosis factor to decrease inflammation are often used to treat autoimmune diseases and inflammatory bowel disease. * Irritable bowel syndrome A
chronic condition, which affects the large intestine and causes abdominal pain, cramping and shifts in bowel movement patterns. In contrast to inflammatory bowel disease, irritable bowel
syndrome is not associated with mucosal inflammation, ulcers or other damage to the bowel. * Chromogranins and secretogranins A family of water-soluble acidic glycoproteins that are mainly
produced by endocrine cells, such as the enteroendocrine cells of the gut. Also known as granins, they are precursors of biologically active peptides involved in inflammation. * Metabolome
All of the metabolites in an environment. * Probiotic An organism or multiple organisms that confer beneficial effects to the host. * Postbiotic A bacterial metabolic product that mediates
benefits to the host. * Prebiotic A certain food or food component that confers a beneficial effect by providing a competitive advantage to beneficial commensal bacteria capable of
metabolizing these substrates or by augmenting the production of metabolic products that result from their fermentation. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Schirmer, M., Garner, A., Vlamakis, H. _et al._ Microbial genes and pathways in inflammatory bowel disease. _Nat Rev Microbiol_ 17, 497–511 (2019).
https://doi.org/10.1038/s41579-019-0213-6 Download citation * Published: 27 June 2019 * Issue Date: August 2019 * DOI: https://doi.org/10.1038/s41579-019-0213-6 SHARE THIS ARTICLE Anyone you
share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the
Springer Nature SharedIt content-sharing initiative
