A male germ-cell-specific ribosome controls male fertility
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Ribosomes are highly sophisticated translation machines that have been demonstrated to be heterogeneous in the regulation of protein synthesis1,2. Male germ cell development
involves complex translational regulation during sperm formation3. However, it remains unclear whether translation during sperm formation is performed by a specific ribosome. Here we report
a ribosome with a specialized nascent polypeptide exit tunnel, RibosomeST, that is assembled with the male germ-cell-specific protein RPL39L, the paralogue of core ribosome (RibosomeCore)
protein RPL39. Deletion of RibosomeST in mice causes defective sperm formation, resulting in substantially reduced fertility. Our comparison of single-particle cryo-electron microscopy
structures of ribosomes from mouse kidneys and testes indicates that RibosomeST features a ribosomal polypeptide exit tunnel of distinct size and charge states compared with RibosomeCore.
RibosomeST predominantly cotranslationally regulates the folding of a subset of male germ-cell-specific proteins that are essential for the formation of sperm. Moreover, we found that
specialized functions of RibosomeST were not replaceable by RibosomeCore. Taken together, identification of this sperm-specific ribosome should greatly expand our understanding of ribosome
function and tissue-specific regulation of protein expression pattern in mammals. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel
any time Learn more Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS A MOLECULAR NETWORK OF CONSERVED FACTORS KEEPS RIBOSOMES DORMANT IN THE EGG Article 18 January
2023 METTL1-DEPENDENT M7G TRNA MODIFICATION IS ESSENTIAL FOR MAINTAINING SPERMATOGENESIS AND FERTILITY IN _DROSOPHILA MELANOGASTER_ Article Open access 24 September 2024 COUPLED PROTEIN
SYNTHESIS AND RIBOSOME-GUIDED PIRNA PROCESSING ON MRNAS Article Open access 13 October 2021 DATA AVAILABILITY The MS proteomics data have been deposited at the ProteomeXchange Consortium via
the PRIDE under project accessions: PXD020874, PXD020864, PXD020922 and PXD037257. Sequencing data were deposited at the National Center for Biotechnology Information (NCBI) Sequence Read
Archive under BioProject PRJNA657531. The cryo-EM density maps reported in this paper are available at the Electron Microscopy Data Bank under accession codes EMD-30432 (80S ribosome from
mouse kidney) and EMD-30433 (80S ribosome from mouse testis); atomic coordinates are reported at the PDB under accession codes 7CPU and 7CPV, respectively. The human 40S and 60S ribosome
structures used in this study were obtained from PDB 6EK0. Source data are provided with this paper. REFERENCES * Genuth, N. R. & Barna, M. The discovery of ribosome heterogeneity and
its implications for gene regulation and organismal life. _Mol. Cell_ 71, 364–374 (2018). Article CAS Google Scholar * Ghulam, M. M., Catala, M. & Abou Elela, S. Differential
expression of duplicated ribosomal protein genes modifies ribosome composition in response to stress. _Nucleic Acids Res._ 48, 1954–1968 (2020). Article CAS Google Scholar * Wang, M. et
al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. _Cell Stem Cell_ 23, 599–614 (2018). Article CAS Google Scholar * Shi, Z. et
al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. _Mol. Cell_ 67, 71–83 (2017). Article CAS Google Scholar * Gupta, V. & Warner, J. R.
Ribosome-omics of the human ribosome. _RNA_ 20, 1004–1013 (2014). Article CAS Google Scholar * Chaillou, T., Zhang, X. & McCarthy, J. J. Expression of muscle-specific ribosomal
protein L3-like impairs myotube growth. _J. Cell. Physiol._ 231, 1894–902 (2016). Article CAS Google Scholar * Kao, B. R. et al. Knockdown of muscle-specific ribosomal protein L3-like
enhances muscle function in healthy and dystrophic mice. _Nucleic Acid Ther._ 31, 457–464 (2021). Article CAS Google Scholar * O’Leary, M. N. et al. The ribosomal protein Rpl22 controls
ribosome composition by directly repressing expression of its own paralog, Rpl22l1. _PLoS Genet._ 9, e1003708 (2013). Article CAS Google Scholar * Rao, S. et al. RPL22L1 induction in
colorectal cancer is associated with poor prognosis and 5-FU resistance. _PLoS ONE_ 14, e0222392 (2019). Article CAS Google Scholar * Kondrashov, N. et al. Ribosome-mediated specificity
in Hox mRNA translation and vertebrate tissue patterning. _Cell_ 145, 383–397 (2011). Article CAS Google Scholar * Simsek, D. et al. The mammalian ribo-interactome reveals ribosome
functional diversity and heterogeneity. _Cell_ 169, 1051–1065 (2017). Article CAS Google Scholar * Slavov, N., Semrau, S., Airoldi, E., Budnik, B. & van Oudenaarden, A. Differential
stoichiometry among core ribosomal proteins. _Cell Rep._ 13, 865–873 (2015). Article CAS Google Scholar * Jiang, L. et al. RPL10L is required for male meiotic division by compensating for
RPL10 during meiotic sex chromosome inactivation in mice. _Curr. Biol._ 27, 1498–1505 (2017). Article CAS Google Scholar * Gamalinda, M. & Woolford Jr, J. L. Deletion of L4 domains
reveals insights into the importance of ribosomal protein extensions in eukaryotic ribosome assembly. _RNA_ 20, 1725–1731 (2014). Article CAS Google Scholar * Anger, A. M. et al.
Structures of the human and _Drosophila_ 80S ribosome. _Nature_ 497, 80–85 (2013). Article ADS CAS Google Scholar * Young, S. A. et al. CABYR is essential for fibrous sheath integrity
and progressive motility in mouse spermatozoa. _J. Cell Sci._ 129, 4379–4387 (2016). CAS Google Scholar * Yang, K., Adham, I. M., Meinhardt, A. & Hoyer-Fender, S. Ultra-structure of
the sperm head-to-tail linkage complex in the absence of the spermatid-specific LINC component SPAG4. _Histochem. Cell Biol._ 150, 49–59 (2018). Article CAS Google Scholar * Shang, Y. et
al. Essential role for SUN5 in anchoring sperm head to the tail. _eLife_ 6, e28199 (2017). Article Google Scholar * Yuan, S. et al. Spata6 is required for normal assembly of the sperm
connecting piece and tight head-tail conjunction. _Proc. Natl Acad. Sci. USA_ 112, E430–E439 (2015). Article CAS Google Scholar * Wang, Y. Y. et al. Deficiency of the _Tbc1d21_ gene
causes male infertility with morphological abnormalities of the sperm mitochondria and flagellum in mice. _PLoS Genet._ 16, e1009020 (2020). Article CAS Google Scholar * Farias-Rico, J.
A., Goetz, S. K., Marino, J. & von Heijne, G. Mutational analysis of protein folding inside the ribosome exit tunnel. _FEBS Lett._ 591, 155–163 (2017). Article CAS Google Scholar *
Kramer, G., Boehringer, D., Ban, N. & Bukau, B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. _Nat. Struct. Mol. Biol._
16, 589–597 (2009). Article CAS Google Scholar * Cassaignau, A. M. E., Cabrita, L. D. & Christodoulou, J. How does the ribosome fold the proteome. _Annu. Rev. Biochem._ 89, 389–415
(2020). Article CAS Google Scholar * Ott, M., Amunts, A. & Brown, A. Organization and regulation of mitochondrial protein synthesis. _Annu. Rev. Biochem._ 85, 77–101 (2016). Article
CAS Google Scholar * Bhushan, S. et al. α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. _Nat. Struct. Mol. Biol._ 17, 313–317 (2010).
Article CAS Google Scholar * Balchin, D., Hayer-Hartl, M. & Hartl, F. U. In vivo aspects of protein folding and quality control. _Science_ 353, aac4354 (2016). Article Google Scholar
* Li, B. et al. A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA-seq. _Sci. Rep._ 7, 4200 (2017). Article ADS Google Scholar * Wang, D. et al. LYPD4, mouse homolog
of a human acrosome protein, is essential for sperm fertilizing ability and male fertility. _Biol. Reprod._ 102, 1033–1044 (2020). Article Google Scholar * Mordret, E. et al. Systematic
detection of amino acid substitutions in proteomes reveals mechanistic basis of ribosome errors and selection for translation fidelity. _Mol. Cell_ 75, 427–441 (2019). Article CAS Google
Scholar * Zou, Q. et al. Proteostasis regulated by testis-specific ribosomal protein RPL39L maintains mouse spermatogenesis. _iScience_ 24, 103396 (2021). Article ADS CAS Google Scholar
* Petrone, P. M., Snow, C. D., Lucent, D. & Pande, V. S. et al. Side-chain recognition and gating in the ribosome exit tunnel. _Proc. Natl Acad. Sci. USA_ 105, 16549–16554 (2008).
Article ADS CAS Google Scholar * Lu, J. & Deutsch, C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. _J. Mol. Biol._ 384, 73–86 (2008). Article CAS Google
Scholar * Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. _Nature_ 596, 583–589 (2021). Article ADS CAS Google Scholar * Varadi, M. et al. AlphaFold
protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. _Nucleic Acids Res._ 50, D439–D444 (2022). Article CAS Google
Scholar * Buhr, F. et al. Synonymous codons direct cotranslational folding toward different protein conformations. _Mol. Cell_ 61, 341–351 (2016). Article CAS Google Scholar * Simpson,
A. J., Caballero, O. L., Jungbluth, A., Chen, Y. T. & Old, L. J. Cancer/testis antigens, gametogenesis and cancer. _Nat. Rev. Cancer_ 5, 615–625 (2005). Article CAS Google Scholar *
Chen, S. et al. Trimethylamine _N_-oxide binds and activates PERK to promote metabolic dysfunction. _Cell Metab._ 30, 1141–1151 (2019). Article CAS Google Scholar * Walter, P. & Ron,
D. The unfolded protein response: from stress pathway to homeostatic regulation. _Science_ 334, 1081–1086 (2011). Article ADS CAS Google Scholar * Pallares, I. & Ventura, S.
Understanding and predicting protein misfolding and aggregation: insights from proteomics. _Proteomics_ 16, 2570–2581 (2016). * Piatek, R., Bruzdziak, P., Wojciechowski, M., Zalewska-Piatek,
B. & Kur, J. The noncanonical disulfide bond as the important stabilizing element of the immunoglobulin fold of the Dr Fimbrial DraE subunit. _Biochemistry_ 49, 1460–1468 (2010).
Article CAS Google Scholar * Tokuhiro, K. et al. OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. _PLoS Genet._ 5, e1000712 (2009). Article Google Scholar
* Koonin, E. V. Orthologs, paralogs, and evolutionary genomics. _Annu. Rev. Genet._ 39, 309–338 (2005). Article CAS Google Scholar * Komili, S., Farny, N. G., Roth, F. P. & Silver, P.
A. Functional specificity among ribosomal proteins regulates gene expression. _Cell_ 131, 557–571 (2007). Article CAS Google Scholar * Gerst, J. E. Pimp my ribosome: ribosomal protein
paralogs specify translational control. _Trends Genet._ 34, 832–845 (2018). Article CAS Google Scholar * Amirbeigiarab, S. et al. Invariable stoichiometry of ribosomal proteins in mouse
brain tissues with aging. _Proc. Natl Acad. Sci. USA_ 116, 22567–22572 (2019). Article ADS CAS Google Scholar * Rinaldi, V. D. et al. An atlas of cell types in the mouse epididymis and
vas deferens. _eLife_ 9, e55474 (2020). Article CAS Google Scholar * Gur, Y. & Breitbart, H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. _Genes
Dev._ 20, 411–416 (2006). Article CAS Google Scholar * Bryant, J. M., Meyer-Ficca, M. L., Dang, V. M., Berger, S. L. & Meyer, R. G. Separation of spermatogenic cell types using
STA-PUT velocity sedimentation. _J. Vis. Exp._ https://doi.org/10.3791/50648 (2013). * Wang, J. P., Page, D. C. & McCarrey, J. R. Differential expression of sex-linked and autosomal
germ-cell-specific genes during spermatogenesis in the mouse. _Hum. Mol. Genet._ 14, 2911–2918 (2005). Article CAS Google Scholar * Guo, X. et al. Proteomic analysis of male 4C germ cell
proteins involved in mouse meiosis. _Proteomics_ 11, 298–308 (2011). Article CAS Google Scholar * Zheng, W. et al. DDB1 regulates sertoli cell proliferation and testis cord remodeling by
TGFβ pathway. _Genes_ 10, 974 (2019). Article CAS Google Scholar * Hua, R. et al. FBXO47 regulates telomere-inner nuclear envelope integration by stabilizing TRF2 during meiosis. _Nucleic
Acids Res._ 47, 11755–11770 (2019). CAS Google Scholar * Mahadevaiah, S. K., Costa, Y. & Turner, J. M. Using RNA FISH to study gene expression during mammalian meiosis. _Methods Mol.
Biol._ 558, 433–444 (2009). Article CAS Google Scholar * Wang, J. et al. Quantitative phosphoproteomics analysis reveals a key role of insulin growth factor 1 receptor (IGF1R) tyrosine
kinase in human sperm capacitation. _Mol. Cell Proteom._ 14, 1104–1112 (2015). Article CAS Google Scholar * Fan, Y. et al. Phosphoproteomic analysis of neonatal regenerative myocardium
revealed important roles of checkpoint kinase 1 via activating mammalian target of rapamycin C1/ribosomal protein S6 kinase b-1 pathway. _Circulation_ 141, 1554–1569 (2020). Article CAS
Google Scholar * Hao, P., Ren, Y., Dutta, B. & Sze, S. K. Comparative evaluation of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) and high-pH reversed phase
(Hp-RP) chromatography in profiling of rat kidney proteome. _J. Proteom._ 82, 254–262 (2013). Article CAS Google Scholar * Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational
platform for mass spectrometry-based shotgun proteomics. _Nat. Protoc._ 11, 2301–2319 (2016). Article CAS Google Scholar * Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal
patterns and correlations in multidimensional genomic data. _Bioinformatics_ 32, 2847–2849 (2016). Article CAS Google Scholar * Yu, G., Wang, L. G., Han, Y. & He, Q. Y.
clusterProfiler: an R package for comparing biological themes among gene clusters. _OMICS_ 16, 284–287 (2012). Article CAS Google Scholar * Peterson, A. C., Russell, J. D., Bailey, D. J.,
Westphall, M. S. & Coon, J. J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. _Mol. Cell. Proteom._ 11, 1475–1488 (2012).
Article Google Scholar * Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. _Nat. Methods_ 14, 331–332 (2017). Article
CAS Google Scholar * Rohou, A. & N G, CTFFIND4: fast and accurate defocus estimation from electron micrographs. _J. Struct. Biol._ 192, 216–221 (2015). Article Google Scholar *
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. _Acta Crystallogr. D_ 73, 496–502 (2017). Article CAS Google Scholar * Zivanov, J.
et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. _eLife_ 7, e42166 (2018). Article Google Scholar * Rosenthal, P. B. & Henderson, R. Optimal
determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. _J. Mol. Biol._ 333, 721–745 (2003). Article CAS Google Scholar *
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. _Nat. Methods_ 11, 63–65 (2014). Article CAS Google Scholar * Emsley, P.,
Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. _Acta Crystallogr. D_ 66, 486–501 (2010). Article CAS Google Scholar * Natchiar, S. K., Myasnikov, A. G.,
Kratzat, H., Hazemann, I. & Klaholz, B. P. Visualization of chemical modifications in the human 80S ribosome structure. _Nature_ 551, 472–477 (2017). Article ADS CAS Google Scholar *
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. _Acta Crystallogr. D_ 75, 861–877 (2019). Article CAS
Google Scholar * Kopylova, E., Noe, L. & Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. _Bioinformatics_ 28, 3211–3217 (2012). Article
CAS Google Scholar * Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. _Nat. Methods_ 12, 357–360 (2015). Article CAS Google Scholar
* Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. _Bioinformatics_ 30, 923–930 (2014). Article
CAS Google Scholar * Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. _Genome Biol._ 15, 550 (2014). Article Google
Scholar * Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. _Bioinformatics_ 26,
139–140 (2010). Article CAS Google Scholar * Tokumoto, S. et al. Development of a Tet-on inducible expression system for the anhydrobiotic cell line, Pv11. _Insects_ 11, 781 (2020).
Article Google Scholar * Hao Zeng, W. X. Ctr9, a key subunit of PAFc, affects global estrogen signaling and drives ERα-positive breast tumorigenesis. _Genes Dev._ 29, 2153–2167 (2015).
Article Google Scholar * Mauthe, M. et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. _Autophagy_ 14, 1435–1455 (2018). Article CAS Google Scholar
* Go, Y. M. & Jones, D. P. Thioredoxin redox western analysis. _Curr. Protoc. Toxicol._ 41, 17.12.1–17.12.12 (2009). * Manning, M. & Colón, W. Structural basis of protein kinetic
stability: resistance to sodium dodecyl sulfate suggests a central role for rigidity and a bias toward beta-sheet structure. _Biochemistry_ 43, 11248–11254 (2004). Article CAS Google
Scholar * Piatek, R. et al. Molecular aspects of biogenesis of _Escherichia coli_ Dr Fimbriae: characterization of DraB-DraE complexes. _Infect. Immun._ 73, 135–145 (2005). Article CAS
Google Scholar * Hu, M. L. Measurement of protein thiol groups and glutathione in plasma. _Methods Enzymol._ 233, 380–385 (1994). Download references ACKNOWLEDGEMENTS We thank D. Li (Center
for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS)) and S. Qian (University of Cornell) for the assistance during the experimental design; B. Zhu, X. Huang and G.
Ji (Institute of Biophysics (IBP), CAS) for cryo-EM data collection at the Center for Biological Imaging (CBI, http://cbi.ibp.ac.cn), IBP, CAS; D. Fan, L. Zhang and B. Huangfu (IBP, CAS) for
assistance during cryo-EM sample screening; Q. Dong (IBP, CAS) for artwork preparation; W. Li (Institute of Zoology, CAS) for assistance in sperm fluorescence in situ hybridization
analyses; and S. Eckardt for help with paper preparation. This work was supported by the National Key R&D Program (2018YFC1003500, 2018YFA0106900 and 2021YFC2700200) and the National
Natural Science Foundation of China (31890784, 31830043). AUTHOR INFORMATION Author notes * These authors contributed equally: Huiling Li, Yangao Huo, Xi He, Liping Yao, Hao Zhang, Yiqiang
Cui, Huijuan Xiao AUTHORS AND AFFILIATIONS * State Key Laboratory of Reproductive Medicine, Department of Histology and Embryology, Nanjing Medical University, Nanjing, China Huiling Li, Xi
He, Liping Yao, Yiqiang Cui, Huijuan Xiao, Wenxiu Xie, Yue Wang, Shu Zhang, Haixia Tu, Yiwei Cheng, Yueshuai Guo, Yunfei Zhu & Xuejiang Guo * CAS Center for Excellence in
Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China Yangao Huo, Dejiu Zhang, Xintao Cao, Tao Jiang & Yan Qin * School of Life Science and Engineering,
Foshan University, Guangdong, China Yangao Huo * Gusu School, Nanjing Medical University, Nanjing, China Hao Zhang * University of Chinese Academy of Sciences, Beijing, China Dejiu Zhang,
Xintao Cao, Tao Jiang & Yan Qin * State Key Laboratory of Reproductive Medicine, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing
Medical University, Nanjing, China Jiahao Sha Authors * Huiling Li View author publications You can also search for this author inPubMed Google Scholar * Yangao Huo View author publications
You can also search for this author inPubMed Google Scholar * Xi He View author publications You can also search for this author inPubMed Google Scholar * Liping Yao View author
publications You can also search for this author inPubMed Google Scholar * Hao Zhang View author publications You can also search for this author inPubMed Google Scholar * Yiqiang Cui View
author publications You can also search for this author inPubMed Google Scholar * Huijuan Xiao View author publications You can also search for this author inPubMed Google Scholar * Wenxiu
Xie View author publications You can also search for this author inPubMed Google Scholar * Dejiu Zhang View author publications You can also search for this author inPubMed Google Scholar *
Yue Wang View author publications You can also search for this author inPubMed Google Scholar * Shu Zhang View author publications You can also search for this author inPubMed Google Scholar
* Haixia Tu View author publications You can also search for this author inPubMed Google Scholar * Yiwei Cheng View author publications You can also search for this author inPubMed Google
Scholar * Yueshuai Guo View author publications You can also search for this author inPubMed Google Scholar * Xintao Cao View author publications You can also search for this author inPubMed
Google Scholar * Yunfei Zhu View author publications You can also search for this author inPubMed Google Scholar * Tao Jiang View author publications You can also search for this author
inPubMed Google Scholar * Xuejiang Guo View author publications You can also search for this author inPubMed Google Scholar * Yan Qin View author publications You can also search for this
author inPubMed Google Scholar * Jiahao Sha View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS H.L., Y.H., X.H., L.Y., H.Z., Y. Cui and H.X.
conducted majority of experiments and analysed the data. H.L., X.H., Y. Cui, H.X., W.X., S.Z., H.T. and Y.Z. performed functional studies in mouse and cell lines. H.L., Y.H., X.H., D.Z.,
X.C. and T.J. performed ribosomal and structural analyses. L.Y., Y.W., Y. Cheng and Y.G. performed proteomics analysis. H.Z. performed bioinformatics analysis. J.S., Y.Q. and X.G. initiated
the project, designed the experiments, supervised the whole project and wrote the manuscript. CORRESPONDING AUTHORS Correspondence to Xuejiang Guo, Yan Qin or Jiahao Sha. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature_ thanks Jon Oatley, Petra Van Damme and the other, anonymous, reviewer(s) for
their contribution to the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and
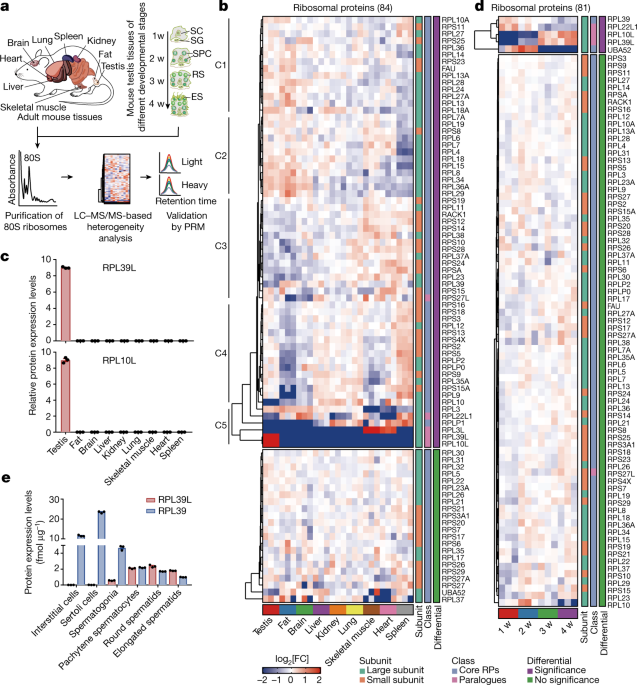
institutional affiliations. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIG. 1 CELL TYPE SPECIFIC RIBOSOME: RIBOSOMAL PROTEOME PROFILES AND RP IDENTIFICATION FROM 80S MONOSOMES ACROSS
MOUSE TISSUES AND DIFFERENT STAGES OF POSTNATAL DEVELOPMENT TESTIS. A, Separation of cytoplasmic ribosomes through sucrose gradient fractionation using different adult mouse tissues. B, Venn
diagram of the identified RPs in a comparative analysis of this and earlier studies. C, Distribution of RP paralogues in different mouse tissues identified in this study. D, Correlation
matrix of cytoplasmic ribosomes from different mouse tissues. E, Density plot of maximal SPM of RPs of nine mouse tissues. F, The relationship between retrogene _Rpl39l_ and its X-linked
parental gene _Rpl39_. G, mRNA levels of _Rpl39_ and _Rpl39l_ in 15 mouse tissues. _n_ = 3. H, Relative quantification of RPL3L in nine mouse tissues using a PRM method. _n_ = 3. I,
Separation of cytoplasmic ribosomes through sucrose gradient fractionation using mouse testicular tissue at different postnatal developmental stages. J, Correlation matrix for cytoplasmic
ribosomes from mouse testicular tissue at different developmental stages. K, Relative protein quantification of RPL4 and RPL39L in non-ribosome, 40S, 60S and ribosome fractions by PRM. _n_ =
3. L, Morphology and specific staining of mouse testicular cells: testicular interstitial cells (IC), Sertoli cells (SC), spermatogonia (SG), pachytene spermatocytes (PS), round spermatids
(RS), and elongated spermatids (ES). Scale bar, 20 μm. M, Purities of SG (82.60 %), PS (82.69 %), RS (87.76 %), ES (81.9 %), IC (92.63 %) and SC (96.35 %) cells. _n_ = 3. N, Quantification
of _Rpl39_ and _Rpl39l_ mRNA levels in testicular cells by qPCR. _n_ = 3. O, Absolute protein quantification of RPL10, RPL10L, and RPL22, RPL22L1 in testicular cells by PRM. _n_ = 3. For G,
H, K, M, N and O, data are mean ± s.e.m. _n_ values represent the number of samples. Source data EXTENDED DATA FIG. 2 THE CRISPR-CAS9 KNOCKOUT STRATEGY AND PHENOTYPE OF RIBOSOMEST-/- MICE.
A, Schematic of CRISPR-Cas9 and sgRNAs used. B, Sequences of the three mutant strains generated by CRISPR-Cas9. C,D, Relative protein expression levels of RPL39L (pachytene spermatocytes:
_P_ = 5.63 × 10−6, spermatids: _P_ = 1.47 × 10−6) (C) and RPL39 (pachytene spermatocytes: _P_ = 1.04 × 10−4, spermatids: _P_ = 1.09 × 10−4) (D) in RibosomeST-/- and wild-type mouse pachytene
spermatocytes and spermatids, as evaluated by PRM. _n_ = 3. E, RibosomeST-/- and wild-type testes with the 40S (_P_ = 0.664), 60S (_P_ = 0.664), 80S (_P_ = 1.25 × 10−6), 2-6 (_P_ = 0.712)
and 7+ polysome (_P_ = 0.489) peaks annotated and statistically compared. _n_ = 5. F, Testis morphology and size of both RibosomeST-/- and wild-type mice. _n_ = 3. Scale bar, 3 mm. G, Sperm
morphology and prevalence of head (_P_ = 7.86 × 10−4) and tail (_P_ = 9.02 × 10−6) defects in RibosomeST-/- and wild-type sperm. _n_ = 4. Scale bar, 10 μm. H, Levels of testosterone (_P_ =
0.662), luteinizing hormone (LH) (_P_ = 0.491) and follicle-stimulating hormone (FSH) (_P_ = 0.437) in RibosomeST-/- and wild-type mice. _n_ = 3. I, Average frequency of vaginal plugs per
female mouse for RibosomeST-/- or wild-type male mice during fertility tests (_P_ = 0.902, _n_ = 3). J, Histology of wild-type and RibosomeST-/- testis showing seminiferous tubules at stage
VII-VIII, circled by dashed lines, and amplified in the bottom panels. _n_ = 3. PL: preleptotene spermatocyte, black arrowhead; PS: pachytene spermatocyte, red arrowhead; RS: round
spermatid, green arrowhead; ES: elongated spermatid, blue arrowhead; SC: Sertoli cell. Top panels scale bar, 50 μm; Bottom panels scale bar, 10 μm. K, Histology of wild-type and
RibosomeST-/- testis showing complete testicular section and stage IX sections together with statistics of percentages of stage IX (_P_ = 0.179) and average number of unreleased sperms per
stage IX tubule (_P_ = 5.87 × 10−4). The red dotted circle indicates the seminiferous tubules of stage IX, and the black dotted line indicates delay-released sperms. Left panels scale bar,
500 μm; Right panels scale bar, 20 μm. _n_ = 3. L, Histology of wild-type and RibosomeST-/- testis showing unreleased sperms at stage IX. Areas marked by dashed lines are shown at high
magnification to the bottom. _n_ = 3. L: leptotene spermatocyte, black arrowhead; PS: pachytene spermatocyte, red arrowhead; ES: elongated spermatid, blue arrowhead; SC: Sertoli cell; RB:
residual body. Top panels scale bar, 50 μm; Bottom panels scale bar, 10 μm. M, The images of testes and testis/body weight ratio (_P_ = 5.63 × 10−4, _n_ = 6) of the three RibosomeST-/-
strains and wild-type mice. Scale bar, 3 mm. N, Epididymal sperm count (Strain 2: _P_ = 0.048, Strain 3: _P_ = 0.045), motility (Strain 2: _P_ = 4.03 × 10−4, Strain 3: _P_ = 4.14 × 10−4) and
progressive motility (Strain 2: _P_ = 3.77 × 10−4, Strain 3: _P_ = 4.05 × 10−4) in two more RibosomeST-/- strains and wild-type mice. _n_ = 3. O, HE staining of the testis and epidydimal
sperms from the three RibosomeST-/- strains. _n_ = 3. Scale bar, 10 μm. P, Fertility of female RibosomeST-/- mice when mated with wild-type male mice (_P_ = 0.606, _n_ = 3). For C, D, E, G,
H, I, K, M, N and P, data are mean ± s.e.m. _P_ values were determined using two-tailed Student’s _t_-tests. _n_ values represent the number of biologically independent animals. Source data
EXTENDED DATA FIG. 3 THE PHENOTYPIC ANALYSIS OF RIBOSOMEST-/- MICE. A, Immunofluorescence detection of LIN28 (red) with nuclei stained by Hoechst 33342 (blue), and the quantification of
positively stained cells in three RibosomeST-/- strains and wild-type mice (Strain 1: _P_ = 0.144, _n_ = 4) (Strain 2: _P_ = 0.759, Strain 3: _P_ = 0.320, _n_ = 3). Left panels scale bar,
500 μm; Right panels scale bar, 20 μm. B, Immunofluorescence detection of SYCP3 (green) and γH2AX (red) in leptotene, zygotene, pachytene and diplotene spermatocytes in RibosomeST-/- and
wild-type testis and the quantification of each stage in RibosomeST-/- and wild-type mice. Scale bar, 10 μm. C, Immunofluorescence detection of SOX9 (red) with nuclei stained by Hoechst
33342 (blue), and the quantification of positively stained cells in RibosomeST-/- and wild-type mice (_P_ = 0.161, _n_ = 3). Left panels scale bar, 50 μm; Right panels scale bar, 20 μm. D,
Fluorescence in situ hybridization of RibosomeST-/- and wild-type sperms using probes specific to X and Y chromosomes, and their corresponding ratio bar plot. Scale bar, 2 μm. E,F,
Transmission electron microscopy analysis and schematic of abnormalities of RibosomeST-/- and wild-type sperms in epididymis. Red arrowhead: defects in doublet microtubules 4-7 of axoneme,
blue arrowhead: other defects of axoneme. Scale bar, 1 μm. (E); The percentage of doublet microtubules 4-7 defects and other defects in axoneme of RibosomeST-/- sperm tail (_P_ = 3.74 ×
10−4, _n_ = 3) (F). G, TUNEL staining (red) of RibosomeST-/- and wild-type testes with Hoechst 33342 stained nuclei (blue), and the quantification statistics of TUNEL positive cells per
tubule (_P_ = 0.009) and percentage of TUNEL positive tubules (_P_ = 0.003, _n_ = 3). Left panels scale bar, 50 μm; Right panels scale bar, 20 μm. For A, C, F and G, data are mean ± s.e.m.
_P_ values were determined using two-tailed Student’s _t_-tests. _n_ values represent the number of biologically independent animals. Source data EXTENDED DATA FIG. 4 PROTEOMICS AND
BIOINFORMATICS ANALYSIS OF DIFFERENTIAL PROTEINS IN RIBOSOMEST-/- SPERMATOCYTES AND ESS. A, Morphology and Hoechst 33342 (blue) staining of spermatocytes and ESs purified by
Fluorescence-Activated Cell Sorting or STA-PUT from RibosomeST-/- and wild-type mice. _n_ = 3. Scale bar, 20 μm. B, The protein expression changes in spermatocytes showing log2 (Fold change)
(RibosomeST-/- vs. wild-type) and –log10 (FDR-adjusted _q_). C, Enriched GO terms for proteins downregulated in RibosomeST-/- ESs. D, Enriched GO terms for proteins upregulated in
RibosomeST-/- ESs. E, Validation of PRM2 (_P_ = 4.06 × 10−5), SMCP (_P_ = 7.70 × 10−6), ATP1A4 (_P_ = 1.23 × 10−5), H1FNT (_P_ = 2.46 × 10−4), SPAG4 (_P_ = 1.41 × 10−5), PGK2 (_P_ = 1,84 ×
10−5), ROPN1 (_P_ = 6.09 × 10−7), ODF1 (_P_ = 1.00 × 10−5), SPATA6 (_P_ = 8.43 × 10−6), TSSK2 (_P_ = 8.22 × 10−5), CST13 (_P_ = 1.80 × 10−6), CRISP2 (_P_ = 2.62 × 10−7), CCIN (_P_ = 5.45 ×
10−5), DBIL5 (_P_ = 4.53 × 10−7), TCP11 (_P_ = 6.34 × 10−9), UBE2J1 (_P_ = 5.30 × 10−7), DCUN1D1 (_P_ = 7.57 × 10−4), and DYNC1LI2 (_P_ = 0.118) in RibosomeST-/- spermatids by relative
targeted quantification using PRM (_n_ = 3). F, Western blot analysis of PRM2 (_P_ = 2.47 × 10−7), PGK2 (_P_ = 0.017), SPATA6 (_P_ = 2.06 × 10−4), TSSK2 (_P_ = 6.99 × 10−4), CRISP2 (_P_ =
0.047), TCP11 (_P_ = 5.92 × 10−4), UBE2J1 (_P_ = 0.007), DCUN1D1 (_P_ = 2.38 × 10−4) and DYNC1LI2 (_P_ = 1.00) in RibosomeST-/- spermatids. _n_ = 3. G, Heat map of clustered mRNA and protein
expression patterns of downregulated proteins with testis-specific expression at different stages of germ cell development. In clusters 1 and 2, the mRNA levels peaked in either
spermatocytes or round spermatids, while the protein levels peaked at ESs. For clusters 3 and 4, all mRNA expression levels peaked in ESs, which agrees with the observed protein expression
patterns. H, Distribution of the testis-specific proteins in downregulated and upregulated differentially expressed proteins in RibosomeST-/- ESs. I, Enriched GO terms for downregulated
testis-specific proteins in RibosomeST-/- ESs. J, Overlap in differentially expressed proteins between RibosomeST-/- and control ESs, and differential genes in 80S, 2-6 and 7+ polysomes
between RibosomeST-/- and wild-type testes by RNC-seq. K, Heat map of mRNA levels in the 80S, 2-6 or 7+ polysomes between RibosomeST-/- and wild-type testes by RNC-seq. L, Western blotting
of nascent peptides using anti-Puromycin in spermatocytes (left) and spermatids (right) following _O_-propargyl-puromycin assays, β-Tubulin was used as a control. _n_ = 3. M, Analysis of
_Ccin_, _Crisp2_, _Cst13_, _Dbil5_, _Dcun1d1_, _H1fnt_, _Odf1_, _Pgk2_, _Prm2_, _Ropn1_, _Spata6_, _Tcp11_, _Tssk2_, _Ube2j1_, _Atp1a4_, _Smcp_, _Spag4_ and _Dync1li2_ mRNA levels in all
polysomal fractions. _n_ = 3. N, Translation ratio of differential proteomics in the 80S, 2-6 or 7+ polysomes between RibosomeST-/- and wild-type testes. O, Matrix of substitution
identifications. Each entry in the matrix represents the number of independent substitutions detected for the corresponding (original codon, destination amino acid) pair in the MOPS-complete
dataset. The logarithmic colour bar highlights the dynamic range of detection. Grey squares indicate substitutions from a codon to its cognate amino acid, substitutions from the stop codon,
or substitutions undetectable via our method because they are indistinguishable from one of the PTMs or artifacts in the UniMod (http://www.unimod.org/) database. For E, F and M, data are
mean ± s.e.m. _P_ values were determined using two-tailed Student’s _t_-tests. _n_ values represent the number of samples. Gel source data are provided in Supplementary Fig. 1. Source data
EXTENDED DATA FIG. 5 CRYO-EM DATA PROCESSING AND RESOLUTION EVALUATION OF KIDNEY AND TESTIS RIBOSOMES. A, Multiple sequence alignment of RPL39 and RPL39L from human, mouse, rat, Drosophila,
_C.elegans_, and yeast. B, Representative micrograph of the kidney ribosome. C, Flow chart of the kidney ribosome cryo-EM data processing. D, Local resolution variation of kidney ribosome
final map estimated with ResMap. E, Gold standard Fourier shell correlation (FSC) curves of the kidney ribosome final map showing that the resolution is 2.82 Å at FSC = 0.143. F,
Representative micrograph of the testis ribosome. G, Flow chart of the testis ribosome cryo-EM data processing. H, Local resolution variation of testis ribosome final map estimated with
ResMap. I, Gold standard Fourier shell correlation (FSC) curves of the testis ribosome final map showing that the resolution is 3.03 Å at FSC = 0.143. EXTENDED DATA FIG. 6 STRUCTURE
COMPARISON OF KIDNEY RIBOSOMECORE, TESTIS RIBOSOMECORE AND TESTIS RIBOSOMEST. A, Structure superposition of 60S subunits of kidney RibosomeCore (pink), testis RibosomeCore (grey), and testis
RibosomeST (brick red). 40S ribosomes were omitted for clarity. C, Close view of RPL39 from kidney RibosomeCore (pink), RPL39 from testis RibosomeCore (grey), and RPL39L from testis
RibosomeST (brick red). B,D, Density of RPL39 from kidney ribosome and density of RPL39 and RPL39L from testis ribosome (level 1.4). F, Close view of sidechains of R28 and R36 of RPL39 from
kidney RibosomeCore (pink), R28 and R36 of RPL39 from testis RibosomeCore (grey),and Q28 and M36 of RPL39L from testis RibosomeST (brick red), indicating orientation of the sidechains. E,G,
Close view of the density of R28 and R36 of RPL39 from kidney RibosomeCore (E); the density of R28 and R36 of RPL39 from testis RibosomeCore, and Q28 and M36 of RPL39L from testis RibosomeST
(G). H,Close view of R28 and R36 of RPL39 from kidney RibosomeCore (pink), with density map omitted for clarity. I,J, Close view of R28 and R36 of RPL39 from testis RibosomeCore (grey; I),
and Q28 and M36 of RPL39L from testis RibosomeST (brick red; J), with density map omitted for clarity. EXTENDED DATA FIG. 7 STRUCTURAL CHARACTERISTIC ANALYSIS OF PROTEINS DOWNREGULATED IN
RIBOSOMEST-/- ESS. A, Downregulated proteins exhibited higher percentage of helix at amino acid positions of 10-11, 14-20, and 23-27 compared with unchanged proteins. Upregulated proteins
exhibited a lower percentage of helix at amino acid positions from 23 to 38 compared with unchanged proteins. B, 3D structure of CCIN based on the AlphaFold Protein Structure Database
(https://alphafold.ebi.ac.uk) with N-terminal 50 amino acids shown in blue, and the C-terminal half sequence shown in grey. C, Downregulated proteins formed more disulfide bonds in
C-terminal half of the sequence than non-significance and upregulated proteins. D, 3D structure of CST13 based on the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk) with
N-terminal 50 amino acids coloured in blue, the C-terminal half sequence coloured in grey, and disulfide bonds are emphasized as red sticks. E, Amino acid enrichment analysis of upregulated
proteins in RibosomeST-/- ESs. F, Levels of total thiols (_P_ = 0.547) and glutathione (GSH) (_P_ = 2.27 × 10−4) in RibosomeST-/- and wild-type N2a cell lines. _n_ = 3. For A and F, data are
mean ± s.e.m. _P_ values were determined using two-sided Fisher’s exact tests for A; two-tailed Student’s _t_-tests for F. _ n_ values represent the number of independent experiments.
Source data EXTENDED DATA FIG. 8 THE CONSTRUCTION OF RIBOSOMEST KNOCKOUT N2A CELL LINE. A, Schematics of CRISPR-Cas9 and sgRNA against _Rpl39l_ used in N2a cells. B, Sequences of the
RibosomeST-/- N2a cell line generated by CRISPR-Cas9. C, Absorbance of ribosome profiles in RibosomeST-/- and wild-type N2a cell lines. D, Validation of TCP11 (_P_ = 0.028), DCUN1D1 (_P_ =
0.006) and DYNC1LI2 (_P_ = 0.916) in RibosomeST-/- N2a cells by western blotting. _n_ = 3. E, Absolute quantification of RPL39 and RPL39L protein expression levels in GC-1 ribosomes, and in
RibosomeST-/- and wild-type N2a ribosomes by PRM (_P_ = 0.004, _n_ = 3). F, Quantification of _Rpl39_ mRNA in RibosomeST-/- and wild-type N2a cells (_P_ = 0.034, _n_ = 3). G, Western
blotting of nascent peptides using anti-Puromycin in N2a cell lines following _O_-propargyl-puromycin assays, β-Tubulin was used as a control. _n_ = 3. H, Quantification of _Prm2_, _H1fnt_,
_Ccin_, and _Dync1li2_ in cell lines and testis. _n_ = 3. For D, E, F and H, data are mean ± s.e.m. _P_ values were determined using two-tailed Student’s _t_-tests. _n_ values represent the
number of independent experiments. Gel source data are provided in Supplementary Fig. 1. Source data EXTENDED DATA FIG. 9 FUNCTIONAL VALIDATION OF RIBOSOMEST IN SPERMATIDS AND CELL LINES. A,
Stability of PRM2 (_P_ = 3.39 × 10−4) and DYNC1LI2 (_P_ = 0.863) in RibosomeST-/- and wild-type spermatids by cycloheximide chase analysis. Protein level of each protein was normalized to
β-Actin and then to time point = 0 h. _n_ = 3. B, Stability of H1FNT-HA in RibosomeST-/- and wild-type N2a cells by cycloheximide chase analysis with the treatment of MG-132. Protein level
was firstly normalized to eGFP and then to time point = 0 h (_P_ = 0.049, _n_ = 3). C, Western blotting showing the relative LC3II/LC3I ratio in RibosomeST-/- and wild-type N2a cells without
(left, _P_ = 0.100, _n_ = 3) or with (right, _P_ = 0.002, _n_ = 5) H1FNT transfection. Stability of H1FNT-HA in RibosomeST-/- and wild-type N2a cells by cycloheximide chase analysis with
the treatment of chloroquine (CQ). Protein level was normalized to eGFP and then to time point = 0 h (_P_ = 0.818, _n_ = 3). D, Stability of PRM2-HA (R28Q: _P_ = 0.021, RPL39L: _P_ = 0.004),
H1FNT-HA (R28Q: _P_ = 0.006, RPL39L: _P_ = 0.001), CCIN-HA (R28Q: _P_ = 0.044, RPL39L: _P_ = 0.021), and DYNC1LI2-HA (R28Q: _P_ = 0.208, RPL39L: _P_ = 0.362) in RibosomeST-/- N2a cell lines
overexpressing RPL39, RPL39(R28Q) and RPL39L by cycloheximide chase analysis, respectively. Protein level of each protein was normalized to eGFP and then to time point = 0 h. _n_ = 3. E,
Unfolded protein response signalling pathway analysis by western blotting of PERK (_P_ = 0.804), eIF2α (_P_ = 0.619), phosphorylated eIF2α (_P_ = 0.608) and ATF6α (_P_ = 2.07 × 10−4) with
β-Tubulin as loading control, qPCR of _Xbp1_ (_Xbp1s_: _P_ = 0.790, _Xbp1u_: _P_ = 0.437, _n_ = 4) splicing with 18S as an internal standard, and relative protein expression levels of
HSP90B1 (FDR- adjusted _q_ = 0.002, fold change < 1.5, Student’s _t_-test with BH FDR correction) and HSPA5 (FDR- adjusted _q_ = 0.019, fold change < 1.5, Student’s _t_-test with BH
FDR correction) according to TMT-based quantification by LC-MS/MS (Supplementary Table 2) in wild-type and RibosomeST-/-spermatids. _n_ = 3. F, Distribution of endogenous proteins MOV10,
GAPDH and β-Tubulin in the soluble (_S_) and pellet (_P_) fractions by western blotting in RibosomeST-/- and wild-type N2a cells. _n_ = 3. G, Distribution of H1FNT-HA (R28Q: _P_ = 0.020,
RPL39L: _P_ = 0.013) and DYNC1LI2-HA (R28Q: _P_ = 0.393, RPL39L: _P_ = 0.465) in the _S_ and _P_ fractions by western blotting and the protein expression levels shown as _S_/_P_ ratio in
RibosomeST-/- N2a cell lines overexpressing RPL39, RPL39(R28Q) and RPL39L, respectively. _n_ = 3. H, SDS-resistance assay for protein OAZ3 synthesized in RibosomeST-/- and wild-type N2a
cells. _n_ = 3. I, Proportion of different wild-type and mutated alleles in wild-type N2a cells, 1st round of Cas9-edited N2a cells, monoclonal heterozygous _Rpl39__+/-_ N2a cell line
derived from 1st round of edit, _Rpl39__+/-_ N2a cells subjected to 2nd round of Cas9-edit and selection for 4 days by blasticidin and puromycin, with and without additional 7 days of
culture without selection. Those without additional 7 days of culture were also subjected to single cell sorting to establish monoclonal cell lines. _n_ = 3. J, Morphology of
RPL39L-IRES-eGFP and doxycycline (Dox)-inducible RPL39-overexpressing RibosomeCore-/Y GC-1 cells with or without Dox treatment for different ranges of time. The number of cells was corrected
according the split ratio during passage. Scale bar, 20 μm. K, Absolute quantification of RPL39 and RPL39L levels in ribosomes of GC-1 wild-type cell, and RPL39L-IRES-eGFP-overexpressing
RibosomeCore-/Y GC-1 cells with or without Dox-induced expression of RPL39 by PRM. _n_ = 3. For A, B, C, D, E, G, I and K, data are mean ± s.e.m. _P_ values were determined using two-tailed
Student’s _t_-tests. _n_ values represent the number of independent experiments. Gel source data are provided in Supplementary Fig. 1. Source data EXTENDED DATA FIG. 10 LINEARITY BETWEEN
THE OBSERVED AND EXPECTED SIGNAL RATIOS OF THE LIGHT AND HEAVY PEPTIDES BY PRM IN SERIAL DILUTION EXPERIMENTS. The observed and expected signal ratios of the light and heavy peptides by PRM
in serial dilution experiments showed good linearity. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Fig. 1 and full descriptions for Supplementary Tables 1–10. REPORTING
SUMMARY SUPPLEMENTARY TABLE 1 Quantification of RPs in the 80S monosomes. SUPPLEMENTARY TABLE 2 Differential proteins observed in different RibosomeST-knockout mouse germ cells.
SUPPLEMENTARY TABLE 3 GO analysis in this study. SUPPLEMENTARY TABLE 4 mRNA and protein expression levels of testis-specific genes in different stages of germ cell development. SUPPLEMENTARY
TABLE 5 RNC-mRNA expression levels in mouse RibosomeST-knockout testis. SUPPLEMENTARY TABLE 6 Cryo-EM data collection and refinement statistics. SUPPLEMENTARY TABLE 7 Subunit lists modelled
in kidney and testis ribosomes. SUPPLEMENTARY TABLE 8 Structural feature analysis of downregulated proteins in mouse RibosomeST-knockout spermatids. SUPPLEMENTARY TABLE 9 Primers and the
peptide sequences used in this study. SUPPLEMENTARY TABLE 10 Extracted ion chromatograms from protein quantification measurements by PRM. SOURCE DATA SOURCE DATA FIG. 1 SOURCE DATA FIG. 2
SOURCE DATA FIG. 4 SOURCE DATA EXTENDED DATA FIG. 1 SOURCE DATA EXTENDED DATA FIG. 2 SOURCE DATA EXTENDED DATA FIG. 3 SOURCE DATA EXTENDED DATA FIG. 4 SOURCE DATA EXTENDED DATA FIG. 7 SOURCE
DATA EXTENDED DATA FIG. 8 SOURCE DATA EXTENDED DATA FIG. 9 RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article
under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such
publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Li, H., Huo, Y., He, X. _et al._ A male germ-cell-specific ribosome controls male
fertility. _Nature_ 612, 725–731 (2022). https://doi.org/10.1038/s41586-022-05508-0 Download citation * Received: 06 September 2021 * Accepted: 01 November 2022 * Published: 14 December 2022
* Issue Date: 22 December 2022 * DOI: https://doi.org/10.1038/s41586-022-05508-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable
link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
