A comprehensive library of human transcription factors for cell fate engineering
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

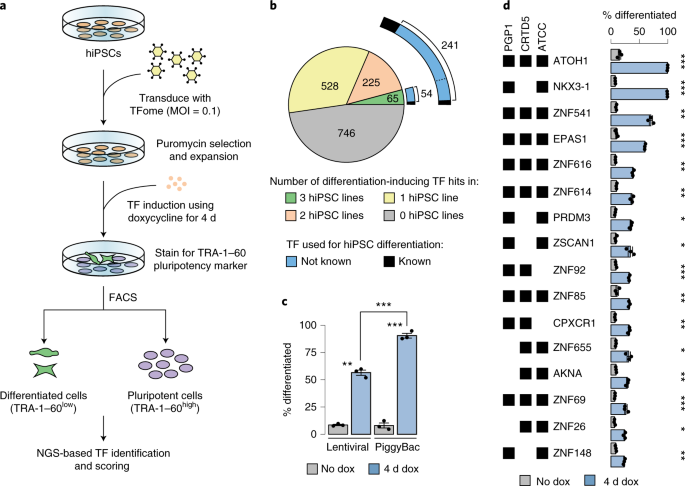
ABSTRACT Human pluripotent stem cells (hPSCs) offer an unprecedented opportunity to model diverse cell types and tissues. To enable systematic exploration of the programming landscape
mediated by transcription factors (TFs), we present the Human TFome, a comprehensive library containing 1,564 TF genes and 1,732 TF splice isoforms. By screening the library in three hPSC
lines, we discovered 290 TFs, including 241 that were previously unreported, that induce differentiation in 4 days without alteration of external soluble or biomechanical cues. We used four
of the hits to program hPSCs into neurons, fibroblasts, oligodendrocytes and vascular endothelial-like cells that have molecular and functional similarity to primary cells. Our
cell-autonomous approach enabled parallel programming of hPSCs into multiple cell types simultaneously. We also demonstrated orthogonal programming by including oligodendrocyte-inducible
hPSCs with unmodified hPSCs to generate cerebral organoids, which expedited in situ myelination. Large-scale combinatorial screening of the Human TFome will complement other strategies for
cell engineering based on developmental biology and computational systems biology. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days
cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS EFFICIENT GENERATION OF FUNCTIONAL NEURONS FROM MOUSE EMBRYONIC STEM CELLS VIA NEUROGENIN-2
EXPRESSION Article 18 August 2023 IDENTIFICATION OF ASCL1 AS A DETERMINANT FOR HUMAN IPSC-DERIVED DOPAMINERGIC NEURONS Article Open access 15 November 2021 ITERATIVE TRANSCRIPTION FACTOR
SCREENING ENABLES RAPID GENERATION OF MICROGLIA-LIKE CELLS FROM HUMAN IPSC Article Open access 10 June 2025 DATA AVAILABILITY. Next-generation sequencing data that support the findings of
the study are available in the Gene Expression Omnibus using accession code GSE159786. CODE AVAILABILITY The code that supports the findings of this study is available from the corresponding
authors upon reasonable request. REFERENCES * Davis, R. L., Weintraub, H. & Lassar, A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. _Cell_ 51, 987–1000
(1987). Article CAS PubMed Google Scholar * Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors.
_Cell_ 126, 663–676 (2006). Article CAS PubMed Google Scholar * Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. _Neuron_ 78, 785–798
(2013). Article CAS PubMed PubMed Central Google Scholar * Parekh, U. et al. Mapping cellular reprogramming via pooled overexpression screens with paired fitness and single-cell
RNA-sequencing readout. _Cell Syst._ 7, 548–555 (2018). Article CAS PubMed PubMed Central Google Scholar * Tsunemoto, R. et al. Diverse reprogramming codes for neuronal identity.
_Nature_ 557, 375–380 (2018). Article CAS PubMed PubMed Central Google Scholar * Pritsker, M., Ford, N. R., Jenq, H. T. & Lemischka, I. R. Genomewide gain-of-function genetic screen
identifies functionally active genes in mouse embryonic stem cells. _Proc. Natl Acad. Sci. USA_ 103, 6946–6951 (2006). Article CAS PubMed PubMed Central Google Scholar * Theodorou, E.
et al. A high throughput embryonic stem cell screen identifies Oct-2 as a bifunctional regulator of neuronal differentiation. _Genes Dev._ 23, 575–588 (2009). Article CAS PubMed PubMed
Central Google Scholar * Yamamizu, K. et al. Identification of transcription factors for lineage-specific ESC differentiation. _Stem Cell Rep._ 1, 545–559 (2013). Article CAS Google
Scholar * Cahan, P. et al. CellNet: network biology applied to stem cell engineering. _Cell_ 158, 903–915 (2014). Article CAS PubMed PubMed Central Google Scholar * Rackham, O. J. et
al. A predictive computational framework for direct reprogramming between human cell types. _Nat. Genet._ 48, 331–335 (2016). Article CAS PubMed Google Scholar * D’Alessio, A. C. et al.
A systematic approach to identify candidate transcription factors that control cell identity. _Stem Cell Rep._ 5, 763–775 (2015). Article Google Scholar * Lambert, S. A. et al. The human
transcription factors. _Cell_ 175, 598–599 (2018). Article CAS PubMed Google Scholar * Nakatake, Y. et al. Generation and profiling of 2,135 human ESC lines for the systematic analyses
of cell states perturbed by inducing single transcription factors. _Cell Rep._ 31, 107655 (2020). Article CAS PubMed Google Scholar * Vaquerizas, J. M., Kummerfeld, S. K., Teichmann, S.
A. & Luscombe, N. M. A census of human transcription factors: function, expression and evolution. _Nat. Rev. Genet._ 10, 252–263 (2009). Article CAS PubMed Google Scholar * Jolma, A.
et al. DNA-binding specificities of human transcription factors. _Cell_ 152, 327–339 (2013). Article CAS PubMed Google Scholar * Seiler, C. Y. et al. DNASU plasmid and
PSI:Biology-Materials repositories: resources to accelerate biological research. _Nucleic Acids Res._ 42, D1253–D1260 (2014). Article CAS PubMed Google Scholar * Yang, X. et al. A public
genome-scale lentiviral expression library of human ORFs. _Nat. Methods_ 8, 659–661 (2011). Article CAS PubMed PubMed Central Google Scholar * Wiemann, S. et al. The ORFeome
Collaboration: a genome-scale human ORF-clone resource. _Nat. Methods_ 13, 191–192 (2016). Article Google Scholar * Adewumi, O. et al. Characterization of human embryonic stem cell lines
by the International Stem Cell Initiative. _Nat. Biotechnol._ 25, 803–816 (2007). Article CAS PubMed Google Scholar * Busskamp, V. et al. Rapid neurogenesis through transcriptional
activation in human stem cells. _Mol. Syst. Biol._ 10, 760 (2014). Article PubMed PubMed Central Google Scholar * Choi, J. et al. A comparison of genetically matched cell lines reveals
the equivalence of human iPSCs and ESCs. _Nat. Biotechnol._ 33, 1173–1181 (2015). Article CAS PubMed PubMed Central Google Scholar * Cahan, P. & Daley, G. Q. Origins and
implications of pluripotent stem cell variability and heterogeneity. _Nat. Rev. Mol. Cell Biol._ 14, 357–368 (2013). Article CAS PubMed PubMed Central Google Scholar * Chanda, S. et al.
Generation of induced neuronal cells by the single reprogramming factor ASCL1. _Stem Cell Rep._3, 282–296 (2014). Article CAS Google Scholar * Bermingham, N. A. et al. Math1: an
essential gene for the generation of inner ear hair cells. _Science_ 284, 1837–1841 (1999). Article CAS PubMed Google Scholar * Sagal, J. et al. Proneural transcription factor Atoh1
drives highly efficient differentiation of human pluripotent stem cells into dopaminergic neurons. _Stem Cells Transl. Med._ 3, 888–898 (2014). Article CAS PubMed PubMed Central Google
Scholar * Xue, Y. et al. Synthetic mRNAs drive highly efficient iPS cell differentiation to dopaminergic neurons. _Stem Cells Transl. Med._ 8, 112–123 (2019). Article CAS PubMed Google
Scholar * Dutta, A. et al. Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. _Science_ 352, 1576–1580 (2016). Article CAS
PubMed PubMed Central Google Scholar * Mai, T. et al. NKX3-1 is required for induced pluripotent stem cell reprogramming and can replace OCT4 in mouse and human iPSC induction. _Nat. Cell
Biol._ 20, 900–908 (2018). Article CAS PubMed PubMed Central Google Scholar * Radley, A. H. et al. Assessment of engineered cells using CellNet and RNA-seq. _Nat. Protoc._ 12,
1089–1102 (2017). Article PubMed PubMed Central Google Scholar * Liang, C. C., Park, A. Y. & Guan, J. L. In vitro scratch assay: a convenient and inexpensive method for analysis of
cell migration in vitro. _Nat. Protoc._ 2, 329–333 (2007). Article CAS PubMed Google Scholar * Bell, E., Ivarsson, B. & Merrill, C. Production of a tissue-like structure by
contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. _Proc. Natl Acad. Sci. USA_ 76, 1274–1278 (1979). Article CAS PubMed PubMed Central
Google Scholar * Lee, D. et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. _Cell Stem Cell_ 2, 497–507 (2008). Article CAS PubMed
PubMed Central Google Scholar * Baralle, F. E. & Giudice, J. Alternative splicing as a regulator of development and tissue identity. _Nat. Rev. Mol. Cell Biol._ 18, 437–451 (2017).
Article CAS PubMed PubMed Central Google Scholar * Potter, R. F. & Groom, A. C. Capillary diameter and geometry in cardiac and skeletal muscle studied by means of corrosion casts.
_Microvasc. Res._ 25, 68–84 (1983). Article CAS PubMed Google Scholar * Schaum, N. et al. Single-cell transcriptomics of 20 mouse organs creates a _Tabula Muris_. _Nature_ 562, 367–372
(2018). Article PubMed Central Google Scholar * Madhavan, M. et al. Induction of myelinating oligodendrocytes in human cortical spheroids. _Nat. Methods_ 15, 700–706 (2018). Article CAS
PubMed PubMed Central Google Scholar * Marton, R. M. et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. _Nat. Neurosci._ 22, 484–491
(2019). Article CAS PubMed PubMed Central Google Scholar * Garcia-Leon, J. A. et al. SOX10 single transcription factor-based fast and efficient generation of oligodendrocytes from human
pluripotent stem cells. _Stem Cell Rep._ 10, 655–672 (2018). Article CAS Google Scholar * Ehrlich, M. et al. Rapid and efficient generation of oligodendrocytes from human induced
pluripotent stem cells using transcription factors. _Proc. Natl Acad. Sci. USA_ 114, E2243–E2252 (2017). Article CAS PubMed PubMed Central Google Scholar * Sarkar, A. &
Hochedlinger, K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. _Cell Stem Cell_ 12, 15–30 (2013). Article CAS PubMed PubMed Central
Google Scholar * Bi, W., Deng, J. M., Zhang, Z., Behringer, R. R. & de Crombrugghe, B. Sox9 is required for cartilage formation. _Nat. Genet._ 22, 85–89 (1999). Article CAS PubMed
Google Scholar * Canals, I. et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. _Nat. Methods_ 15, 693–696 (2018). Article CAS PubMed Google
Scholar * Khoshakhlagh, P., Sivakumar, A., Pace, L. A., Sazer, D. W. & Moore, M. J. Methods for fabrication and evaluation of a 3D microengineered model of myelinated peripheral nerve.
_J. Neural Eng._ 15, 064001 (2018). Article PubMed PubMed Central Google Scholar * Khoshakhlagh, P. & Moore, M. J. Photoreactive interpenetrating network of hyaluronic acid and
Puramatrix as a selectively tunable scaffold for neurite growth. _Acta Biomater._ 16, 23–34 (2015). Article CAS PubMed Google Scholar * Mohammadi, S. et al. Whole-brain in-vivo
measurements of the axonal G-ratio in a group of 37 healthy volunteers. _Front. Neurosci._ 9, 441 (2015). Article PubMed PubMed Central Google Scholar * Windrem, M. S. et al. Fetal and
adult human oligodendrocyte progenitor cell isolates myelinate the congenitally dysmyelinated brain. _Nat. Med._ 10, 93–97 (2004). Article CAS PubMed Google Scholar * Lancaster, M. A. et
al. Cerebral organoids model human brain development and microcephaly. _Nature_ 501, 373–379 (2013). Article CAS PubMed Google Scholar * Togo, S. et al. Differentiation of embryonic
stem cells into fibroblast-like cells in three-dimensional type I collagen gel cultures. _In Vitro Cell. Dev. Biol. Anim._ 47, 114–124 (2011). Article CAS PubMed Google Scholar *
Elcheva, I. et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. _Nat. Commun._ 5, 4372 (2014). Article CAS PubMed Google
Scholar * Morita, R. et al. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. _Proc. Natl Acad. Sci. USA_ 112, 160–165 (2015). Article
CAS PubMed Google Scholar * Cakir, B. et al. Engineering of human brain organoids with a functional vascular-like system. _Nat. Methods_ 16, 1169–1175 (2019). Article CAS PubMed PubMed
Central Google Scholar * Woltjen, K. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. _Nature_ 458, 766–770 (2009). Article CAS PubMed PubMed
Central Google Scholar * Ronaldson-Bouchard, K. & Vunjak-Novakovic, G. Organs-on-a-Chip: a fast track for engineered human tissues in drug development. _Cell Stem Cell_ 22, 310–324
(2018). Article CAS PubMed PubMed Central Google Scholar * Guye, P. et al. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using
Gata6. _Nat. Commun._ 7, 10243 (2016). Article CAS PubMed PubMed Central Google Scholar * Bagley, J. A., Reumann, D., Bian, S., Levi-Strauss, J. & Knoblich, J. A. Fused cerebral
organoids model interactions between brain regions. _Nat. Methods_ 14, 743–751 (2017). Article CAS PubMed PubMed Central Google Scholar * Birey, F. et al. Assembly of functionally
integrated human forebrain spheroids. _Nature_ 545, 54–59 (2017). Article CAS PubMed PubMed Central Google Scholar * Xiang, Y. et al. Fusion of regionally specified hPSC-derived
organoids models human brain development and interneuron migration. _Cell Stem Cell_ 21, 383–398 (2017). Article CAS PubMed PubMed Central Google Scholar * Cederquist, G. Y. et al.
Specification of positional identity in forebrain organoids. _Nat. Biotechnol._ 37, 436–444 (2019). Article CAS PubMed PubMed Central Google Scholar * Mansour, A. A. et al. An in vivo
model of functional and vascularized human brain organoids. _Nat. Biotechnol._ 36, 432–441 (2018). Article CAS PubMed PubMed Central Google Scholar * Rozenblatt-Rosen, O., Stubbington,
M. J. T., Regev, A. & Teichmann, S. A. The Human Cell Atlas: from vision to reality. _Nature_ 550, 451–453 (2017). Article CAS PubMed Google Scholar * Han, X. et al. Construction of
a human cell landscape at single-cell level. _Nature_ 581, 303–309 (2020). * Cusanovich, D. A. et al. A single-cell atlas of in vivo mammalian chromatin accessibility. _Cell_ 174, 1309–1324
(2018). Article CAS PubMed PubMed Central Google Scholar * Moss, J. et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and
disease. _Nat. Commun._ 9, 5068 (2018). Article PubMed PubMed Central Google Scholar * Gray, K. A., Yates, B., Seal, R. L., Wright, M. W. & Bruford, E. A. Genenames.org: the HGNC
resources in 2015. _Nucleic Acids Res._ 43, D1079–D1085 (2015). Article CAS PubMed Google Scholar * Mele, M. et al. Human genomics. The human transcriptome across tissues and
individuals. _Science_ 348, 660–665 (2015). Article CAS PubMed PubMed Central Google Scholar * Church, G. M. The personal genome project. _Mol. Syst. Biol._ 1, 2005.0030 (2005). Article
CAS PubMed PubMed Central Google Scholar * Kutsche, L. K. et al. Combined experimental and system-level analyses reveal the complex regulatory network of miR-124 during human
neurogenesis. _Cell Syst._ 7, 438–452 (2018). Article CAS PubMed PubMed Central Google Scholar * Salmon, P. & Trono, D. Production and titration of lentiviral vectors. in _Current
Protocols in Human Genetic_s Ch. 12, Unit 12.10 (Wiley, 2007). * Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. _Bioinformatics_ 29, 15–21 (2013). Article CAS PubMed Google
Scholar * Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. _Genome Biol._ 15, 550 (2014). Article PubMed PubMed
Central Google Scholar * Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. _J. Neurosci._ 34, 11929–11947
(2014). Article CAS PubMed PubMed Central Google Scholar * Zhang, J. et al. A genome-wide analysis of human pluripotent stem cell-derived endothelial cells in 2D or 3D culture. _Stem
Cell Rep._ 8, 907–918 (2017). Article CAS Google Scholar * Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. _BMC
Bioinf._ 12, 323 (2011). Article CAS Google Scholar * Bult, C. J., Blake, J. A., Smith, C. L., Kadin, J. A. & Richardson, J. E. Mouse genome database (MGD) 2019. _Nucleic Acids Res._
47, D801–D806 (2019). Article CAS PubMed Google Scholar * Anders, S. & Huber, W. Differential expression analysis for sequence count data. _Genome Biol._ 11, R106 (2010). Article
CAS PubMed PubMed Central Google Scholar * Leek, J. T. svaseq: removing batch effects and other unwanted noise from sequencing data. _Nucleic Acids Res_. 42, e161 (2014). * Schindelin,
J. et al. Fiji: an open-source platform for biological-image analysis. _Nat. Methods_ 9, 676–682 (2012). Article CAS PubMed Google Scholar * Ngo, P., Ramalingam, P., Phillips, J. A.
& Furuta, G. T. Collagen gel contraction assay. _Methods Mol. Biol._ 341, 103–109 (2006). CAS PubMed Google Scholar * Picelli, S. et al. Smart-seq2 for sensitive full-length
transcriptome profiling in single cells. _Nat. Methods_ 10, 1096–1098 (2013). Article CAS PubMed Google Scholar * Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale
single-cell gene expression data analysis. _Genome Biol._ 19, 15 (2018). Article PubMed PubMed Central Google Scholar * Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient
general purpose program for assigning sequence reads to genomic features. _Bioinformatics_ 30, 923–930 (2014). Article CAS PubMed Google Scholar * Koike, N. et al. Tissue engineering:
creation of long-lasting blood vessels. _Nature_ 428, 138–139 (2004). Article CAS PubMed Google Scholar * Melero-Martin, J. M. et al. Engineering robust and functional vascular networks
in vivo with human adult and cord blood-derived progenitor cells. _Circ. Res._ 103, 194–202 (2008). Article CAS PubMed PubMed Central Google Scholar * Khoshakhlagh, P. et al.
Development and characterization of a bioglass/chitosan composite as an injectable bone substitute. _Carbohydrate Polym._ 157, 1261–1271 (2017). Article CAS Google Scholar * Khoshakhlagh,
P., Bowser, D. A., Brown, J. Q. & Moore, M. J. Comparison of visible and UVA phototoxicity in neural culture systems micropatterned with digital projection photolithography. _J. Biomed.
Mater. Res. A_ 107, 134–144 (2019). Article CAS PubMed Google Scholar * Douvaras, P. & Fossati, V. Generation and isolation of oligodendrocyte progenitor cells from human
pluripotent stem cells. _Nat. Protoc._ 10, 1143–1154 (2015). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank J. Aach, M. O. Karl, R. Kalhor, N. Ostrov and
H. Lee for critical feedback and the Church and Busskamp laboratories for support. We acknowledge technical support from the Harvard Biopolymers Facility, the Harvard Division of Immunology
Flow Cytometry Core Facility, the Beth Israel Deaconess Medical Center Flow Cytometry Core, the Wyss Flow Cytometry and Microscopy Core, M. Ericsson and P. Coughlin at the Harvard Medical
School Electron Microscopy Facility, M. T. Gianatasio at the Dana-Farber/Harvard Cancer Center Specialized Histopathology Core and Rodent Histopathology Core (both supported, in part, by
National Cancer Institute Cancer Center Support grant NIH 5 P30 CA06516) and Harvard Medical School Orchestra Research Computing. We also thank the TU Dresden Center for Molecular and
Cellular Bioengineering Advanced Imaging, Deep Sequencing, Flow Cytometry and Stem Cell Engineering core facilities. We would also like to thank J. Gray’s laboratory for electrophysiology
support, S. Jeanty and J. Lee (Church lab, Harvard Medical School) for the PGP1 Sendai virus hiPSC line, G. Sheynkman and W. Glindmeyer for helpful discussions, A. Jolma, K. Nitta and K.
Said for technical assistance and M. Lemieux and J. McDade for their support in depositing the library to Addgene. A.H.M.N. was supported by an NSERC Postgraduate Fellowship and a Peter and
Carolyn Lynch Foundation Fellowship. J.E.R.A. was supported by the DIGS-BB program. S.L.S. is a Shurl and Kay Curci Foundation Fellow of the Life Sciences Research Foundation. The Ellison
Foundation and Institute Sponsored Research funds from the DFCI Strategic Initiative supported M.V. and D.E.H. The project was supported by the Volkswagen Foundation (Freigeist - A110720),
the European Research Council (ERC-StG-678071 - ProNeurons) and the Deutsche Forschungsgemeinschaft (SPP2127, EXC-2068-390729961 - Cluster of Excellence - Physics of Life at TU Dresden and
EXC-2151-390873048 – Cluster of Excellence – ImmunoSensation2 at the University of Bonn) to V.B. G.M.C. acknowledges funding from National Human Genome Research Institute grants P50 HG005550
‘Center for Casual Variation’, RM1 HG008525 ‘Center for Genomically Engineered Organs’, the Simons Foundation for Autism Research Initiative (368485), the Blavatnik Biomedical Accelerator
at Harvard University, the FunGCAT program from the Office of the Director of National Intelligence Intelligence Advanced Research Projects Activity, via the Army Research Office, under
federal award no. W911NF-17-2-0089 and research funding from R. Merkin and the Merkin Family Foundation. AUTHOR INFORMATION Author notes * Jesus Eduardo Rojo Arias Present address:
Wellcome-MRC Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre, Cambridge Biomedical Campus, University of Cambridge, Cambridge, UK * These authors contributed equally: Alex H.
M. Ng, Parastoo Khoshakhlagh. AUTHORS AND AFFILIATIONS * Department of Genetics, Blavatnik Institute, Harvard Medical School, Boston, MA, USA Alex H. M. Ng, Parastoo Khoshakhlagh, Evan
Appleton, Kiavash Kiaee, Richie E. Kohman, Matthew Dysart, Kathleen Leeper, Wren Saylor, Jeremy Y. Huang, David E. Hill, Marc Vidal & George M. Church * Wyss Institute for Biologically
Inspired Engineering at Harvard University, Boston, MA, USA Alex H. M. Ng, Parastoo Khoshakhlagh, Evan Appleton, Kiavash Kiaee, Richie E. Kohman, Andyna Vernet, Matthew Dysart, Kathleen
Leeper, Wren Saylor, Jeremy Y. Huang, Amanda Graveline & George M. Church * GC Therapeutics, Inc, Cambridge, MA, USA Alex H. M. Ng, Parastoo Khoshakhlagh, Evan Appleton, Kiavash Kiaee
& George M. Church * Technische Universität Dresden, Center for Molecular and Cellular Bioengineering (CMCB), Center for Regenerative Therapies Dresden (CRTD), Dresden, Germany Jesus
Eduardo Rojo Arias, Giovanni Pasquini, Anka Swiersy & Volker Busskamp * Department of Cardiac Surgery, Boston Children’s Hospital, Boston, MA, USA Kai Wang & Juan M. Melero-Martin *
Department of Surgery, Harvard Medical School, Boston, MA, USA Kai Wang & Juan M. Melero-Martin * Gladstone Institutes and University of California, San Francisco, San Francisco, CA, USA
Seth L. Shipman * Department of Biochemistry, University of Cambridge, Cambridge, UK Jussi Taipale * Department of Medical Biochemistry and Biophysics, Karolinska Institute, Stockholm,
Sweden Jussi Taipale * Applied Tumor Genomics Program, Faculty of Medicine, University of Helsinki, Helsinki, Finland Jussi Taipale * Center for Cancer Systems Biology (CCSB), Dana-Farber
Cancer Institute, Boston, MA, USA David E. Hill & Marc Vidal * Department of Ophthalmology, Medical Faculty, University of Bonn, Bonn, Germany Volker Busskamp Authors * Alex H. M. Ng
View author publications You can also search for this author inPubMed Google Scholar * Parastoo Khoshakhlagh View author publications You can also search for this author inPubMed Google
Scholar * Jesus Eduardo Rojo Arias View author publications You can also search for this author inPubMed Google Scholar * Giovanni Pasquini View author publications You can also search for
this author inPubMed Google Scholar * Kai Wang View author publications You can also search for this author inPubMed Google Scholar * Anka Swiersy View author publications You can also
search for this author inPubMed Google Scholar * Seth L. Shipman View author publications You can also search for this author inPubMed Google Scholar * Evan Appleton View author publications
You can also search for this author inPubMed Google Scholar * Kiavash Kiaee View author publications You can also search for this author inPubMed Google Scholar * Richie E. Kohman View
author publications You can also search for this author inPubMed Google Scholar * Andyna Vernet View author publications You can also search for this author inPubMed Google Scholar * Matthew
Dysart View author publications You can also search for this author inPubMed Google Scholar * Kathleen Leeper View author publications You can also search for this author inPubMed Google
Scholar * Wren Saylor View author publications You can also search for this author inPubMed Google Scholar * Jeremy Y. Huang View author publications You can also search for this author
inPubMed Google Scholar * Amanda Graveline View author publications You can also search for this author inPubMed Google Scholar * Jussi Taipale View author publications You can also search
for this author inPubMed Google Scholar * David E. Hill View author publications You can also search for this author inPubMed Google Scholar * Marc Vidal View author publications You can
also search for this author inPubMed Google Scholar * Juan M. Melero-Martin View author publications You can also search for this author inPubMed Google Scholar * Volker Busskamp View author
publications You can also search for this author inPubMed Google Scholar * George M. Church View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS A.H.M.N., P.K., V.B. and G.M.C. conceived the idea, led the study and designed all experiments. A.H.M.N. and P.K. performed most of the experiments and analyses, with
significant technical contributions from J.E.R.A, G.P., K.W., A.S., S.L.S., E.A., K.K., R.E.K., A.V., M.D., K.L., W.S., J.Y.H., A.G., J.T., D.E.H., M.V. and J.M.M.-M. V.B. and G.M.C. oversaw
the study. A.H.M.N., P.K. and V.B. wrote the manuscript with input and feedback from all authors. CORRESPONDING AUTHORS Correspondence to Volker Busskamp or George M. Church. ETHICS
DECLARATIONS COMPETING INTERESTS A.H.M.N., P.K., V.B. and G.M.C. are inventors on patents filed by the Presidents and Fellows of Harvard College. Full disclosure for G.M.C. is available at
http://arep.med.harvard.edu/gmc/tech.html. A.H.M.N., P.K. and G.M.C. are co-founders of and have equity in GC Therapeutics, Inc. No reagents or funding from GC Therapeutics were used in this
study. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figs. 1–7 REPORTING SUMMARY SUPPLEMENTARY TABLE 1 TFs in the Human TFome SUPPLEMENTARY TABLE 2 TFome screen sequencing statistics
SUPPLEMENTARY TABLE 3 TFome screen differentiation scores SUPPLEMENTARY TABLE 4 Novelty and tissue expression of 290 TF hits SUPPLEMENTARY TABLE 5 RNA-seq statistics and expression profiles
SUPPLEMENTARY TABLE 6 TFs involved in oligodendrocyte development SUPPLEMENTARY TABLE 7 Exact _P_ values for statistical tests RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS
ARTICLE CITE THIS ARTICLE Ng, A.H.M., Khoshakhlagh, P., Rojo Arias, J.E. _et al._ A comprehensive library of human transcription factors for cell fate engineering. _Nat Biotechnol_ 39,
510–519 (2021). https://doi.org/10.1038/s41587-020-0742-6 Download citation * Received: 03 November 2019 * Accepted: 19 October 2020 * Published: 30 November 2020 * Issue Date: April 2021 *
DOI: https://doi.org/10.1038/s41587-020-0742-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is
not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
