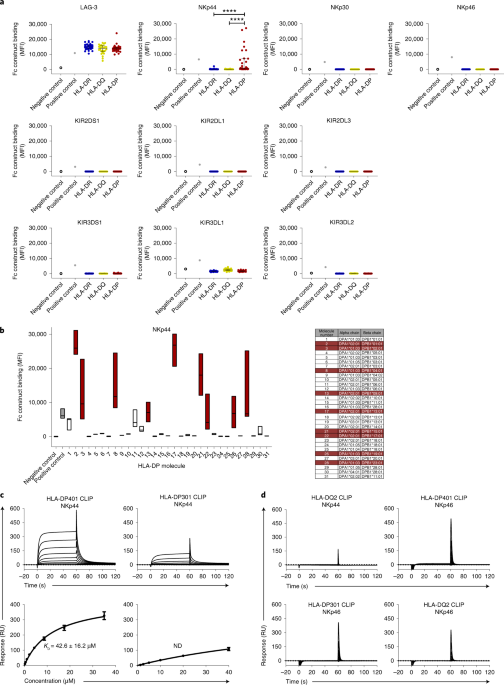
A subset of hla-dp molecules serve as ligands for the natural cytotoxicity receptor nkp44
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Natural killer (NK) cells can recognize virus-infected and stressed cells1 using activating and inhibitory receptors, many of which interact with HLA class I. Although early studies
also suggested a functional impact of HLA class II on NK cell activity2,3, the NK cell receptors that specifically recognize HLA class II molecules have never been identified. We
investigated whether two major families of NK cell receptors, killer-cell immunoglobulin-like receptors (KIRs) and natural cytotoxicity receptors (NCRs), contained receptors that bound to
HLA class II, and identified a direct interaction between the NK cell receptor NKp44 and a subset of HLA-DP molecules, including HLA-DP401, one of the most frequent class II allotypes in
white populations4. Using NKp44ζ+ reporter cells and primary human NKp44+ NK cells, we demonstrated that interactions between NKp44 and HLA-DP401 trigger functional NK cell responses. This
interaction between a subset of HLA-DP molecules and NKp44 implicates HLA class II as a component of the innate immune response, much like HLA class I. It also provides a potential mechanism
for the described associations between HLA-DP subtypes and several disease outcomes, including hepatitis B virus infection5,6,7, graft-versus-host disease8 and inflammatory bowel
disease9,10. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access
Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print
issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to
local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT
BEING VIEWED BY OTHERS TLR9 AGONISM DIFFERENTIALLY IMPACTS HUMAN NK CELL-MEDIATED DIRECT KILLING AND ANTIBODY-DEPENDENT CELL-MEDIATED CYTOTOXICITY Article Open access 25 June 2024 MOUSE AND
HUMAN ANTIBODIES BIND HLA-E-LEADER PEPTIDE COMPLEXES AND ENHANCE NK CELL CYTOTOXICITY Article Open access 28 March 2022 DISTINCT CD16A FEATURES ON HUMAN NK CELLS OBSERVED BY FLOW CYTOMETRY
CORRELATE WITH INCREASED ADCC Article Open access 04 April 2024 DATA AVAILABILITY All figures have associated raw data. All primary data files are available upon request from the
corresponding author. CODE AVAILABILITY All codes are available upon request from the corresponding author REFERENCES * Jost, S. & Altfeld, M. Control of human viral infections by
natural killer cells. _Annu. Rev. Immunol._ 31, 163–194 (2013). Article CAS Google Scholar * Jiang, Y. Z. et al. Interaction of natural killer cells with MHC class II: reversal of
HLA-DR1-mediated protection of K562 transfectant from natural killer cell-mediated cytolysis by brefeldin-A. _Immunology_ 87, 481–486 (1996). Article CAS Google Scholar * Lobo, P. I.,
Chang, M. Y. & Mellins, E. Mechanisms by which HLA-class II molecules protect human B lymphoid tumour cells against NK- and LAK-mediated cytolysis. _Immunology_ 88, 625–629 (1996).
Article CAS Google Scholar * al-Daccak, R. et al. Gene polymorphism of HLA-DPB1 and DPA1 loci in caucasoid population: frequencies and DPB1-DPA1 associations. _Hum. Immunol._ 31, 277–285
(1991). Article CAS Google Scholar * Guo, X. et al. Strong influence of human leukocyte antigen (HLA)-DP gene variants on development of persistent chronic hepatitis B virus carriers in
the Han Chinese population. _Hepatology_ 53, 422–428 (2011). Article CAS Google Scholar * Kamatani, Y. et al. A genome-wide association study identifies variants in the HLA-DP locus
associated with chronic hepatitis B in Asians. _Nat. Genet._ 41, 591–595 (2009). Article CAS Google Scholar * Thomas, R. et al. A novel variant marking HLA-DP expression levels predicts
recovery from hepatitis B virus infection. _J. Virol._ 86, 6979–6985 (2012). Article Google Scholar * Petersdorf, E. W. et al. High HLA-DP expression and graft-versus-host disease. _N.
Engl. J. Med._ 373, 599–609 (2015). Article CAS Google Scholar * Hadley, D. et al. HLA-DPB1*04:01 protects genetically susceptible children from celiac disease autoimmunity in the TEDDY
study. _Am. J. Gastroenterol._ 110, 915–920 (2015). Article CAS Google Scholar * Goyette, P. et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in
inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. _Nat. Genet._ 47, 172–179 (2015). Article CAS Google Scholar * Garcia-Beltran, W. F. et al. Open conformers
of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. _Nat. Immunol._ 17, 1067–1074 (2016). Article CAS Google Scholar * Triebel, F. et al. LAG-3, a novel
lymphocyte activation gene closely related to CD4. _J. Exp. Med._ 171, 1393–1405 (1990). Article CAS Google Scholar * Berry, R. et al. Targeting of a natural killer cell receptor family
by a viral immunoevasin. _Nat. Immunol._ 14, 699–705 (2013). Article CAS Google Scholar * Deuss, F. A., Watson, G. M., Fu, Z., Rossjohn, J. & Berry, R. Structural basis for CD96
immune receptor recognition of nectin-like protein-5, CD155. _Structure_ 27, 219–228.e3 (2019). Article CAS Google Scholar * Rossjohn, J. et al. T cell antigen receptor recognition of
antigen-presenting molecules. _Annu. Rev. Immunol._ 33, 169–200 (2015). Article CAS Google Scholar * Vivian, J. P. et al. Killer cell immunoglobulin-like receptor 3DL1-mediated
recognition of human leukocyte antigen B. _Nature_ 479, 401–405 (2011). Article CAS Google Scholar * Vitale, M. et al. NKp44, a novel triggering surface molecule specifically expressed by
activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. _J. Exp. Med._ 187, 2065–2072 (1998). Article CAS Google Scholar * Alter,
G., Malenfant, J. M. & Altfeld, M. CD107a as a functional marker for the identification of natural killer cell activity. _J. Immunol. Methods_ 294, 15–22 (2004). Article CAS Google
Scholar * Hiltbold, E. M. & Roche, P. A. Trafficking of MHC class II molecules in the late secretory pathway. _Curr. Opin. Immunol._ 14, 30–35 (2002). Article CAS Google Scholar *
Wen, F., Esteban, O. & Zhao, H. Rapid identification of CD4+ T-cell epitopes using yeast displaying pathogen-derived peptide library. _J. Immunol. Methods_ 336, 37–44 (2008). Article
CAS Google Scholar * Birnbaum, M. E. et al. Deconstructing the peptide-MHC specificity of T cell recognition. _Cell_ 157, 1073–1087 (2014). Article CAS Google Scholar * Lo, W.-L. et al.
An endogenous peptide positively selects and augments the activation and survival of peripheral CD4+ T cells. _Nat. Immunol._ 10, 1155–1161 (2009). Article CAS Google Scholar * Dai, S.
et al. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. _Proc. Natl Acad. Sci. USA_ 107, 7425–7430 (2010). Article CAS Google Scholar * Stevanovic, S. et al.
HLA class II upregulation during viral infection leads to HLA-DP-directed graft-versus-host disease after CD4+ donor lymphocyte infusion. _Blood_ 122, 1963–1973 (2013). Article CAS Google
Scholar * Holzemer, A. et al. Selection of an HLA-C*03:04-restricted HIV-1 p24 Gag sequence variant is associated with viral escape from KIR2DL3+ natural killer cells: data from an
observational cohort in South Africa. _PLoS Med._ 12, e1001900 (2015). Article Google Scholar * Naiyer, M. M. et al. KIR2DS2 recognizes conserved peptides derived from viral helicases in
the context of HLA-C. _Sci. Immunol._ 2, eaal5296 (2017). Article Google Scholar * O’Connor, G. M. et al. Peptide-dependent recognition of HLA-B*57:01 by KIR3DS1. _J. Virol._ 89, 5213–5221
(2015). Article Google Scholar * O’Connor, G. M. et al. Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide
sensitivity. _J. Immunol._ 192, 2875–2884 (2014). Article Google Scholar * Chapel, A. et al. Peptide-specific engagement of the activating NK cell receptor KIR2DS1. _Sci. Rep._ 7, 2414
(2017). Article Google Scholar * Rajagopalan, S. & Long, E. O. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4
exhibits peptide selectivity. _J. Exp. Med._ 185, 1523–1528 (1997). Article CAS Google Scholar * Holling, T. M., Schooten, E., Langerak, A. W. & van den Elsen, P. J. Regulation of MHC
class II expression in human T-cell malignancies. _Blood_ 103, 1438–1444 (2004). Article CAS Google Scholar * Thompson, J. A. et al. Tumor cells transduced with the MHC class II
transactivator and CD80 activate tumor-specific CD4+ T cells whether or not they are silenced for invariant chain. _Cancer Res._ 66, 1147–1154 (2006). Article CAS Google Scholar *
Takayama, T. et al. Imbalance of NKp44+NKp46− and NKp44−NKp46+ natural killer cells in the intestinal mucosa of patients with Crohn’s disease. _Gastroenterology_ 139, 882–892.e3 (2010).
Article CAS Google Scholar * Glatzer, T. et al. RORgammat+ innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. _Immunity_ 38,
1223–1235 (2013). Article CAS Google Scholar * Campbell, K. S., Yusa, S., Kikuchi-Maki, A. & Catina, T. L. NKp44 triggers NK cell activation through DAP12 association that is not
influenced by a putative cytoplasmic inhibitory sequence. _J. Immunol._ 172, 899–906 (2004). Article CAS Google Scholar * Cantoni, C. et al. NKp44, a triggering receptor involved in tumor
cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. _J. Exp. Med._ 189, 787–796 (1999). Article CAS Google Scholar * Siewiera, J. et
al. Natural cytotoxicity receptor splice variants orchestrate the distinct functions of human natural killer cell subtypes. _Nat. Commun._ 6, 10183 (2015). Article CAS Google Scholar *
Arnon, T. I. et al. Recognition of viral hemagglutinins by NKp44 but not by NKp30. _Eur. J. Immunol._ 31, 2680–2689 (2001). Article CAS Google Scholar * Ho, J. W. et al. H5-type influenza
virus hemagglutinin is functionally recognized by the natural killer-activating receptor NKp44. _J. Virol._ 82, 2028–2032 (2008). Article CAS Google Scholar * Rosental, B. et al.
Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. _J. Immunol._ 187, 5693–5702 (2011). Article CAS Google Scholar * Baychelier,
F. et al. Identification of a cellular ligand for the natural cytotoxicity receptor NKp44. _Blood_ 122, 2935–2942 (2013). Article CAS Google Scholar * Barrow, A. D. et al. Natural killer
cells control tumor growth by sensing a growth factor. _Cell_ 172, 534–548.e19 (2018). Article CAS Google Scholar * De Maria, A. et al. NKp44 expression, phylogenesis and function in
non-human primate NK cells. _Int. Immunol._ 21, 245–255 (2009). Article Google Scholar * Ting, J. P.-Y. & Trowsdale, J. Genetic control of MHC class II expression. _Cell_ 109, S21–S33
(2002). Article CAS Google Scholar * Allcock, R. J. N., Barrow, A. D., Forbes, S., Beck, S. & Trowsdale, J. The human TREM gene cluster at 6p21.1 encodes both activating and
inhibitory single IgV domain receptors and includes NKp44. _Eur. J. Immunol._ 33, 567–577 (2003). Article CAS Google Scholar * Slierendregt, B. L., Otting, N., Kenter, M. & Bontrop,
R. E. Allelic diversity at the Mhc-DP locus in rhesus macaques (_Macaca mulatta_). _Immunogenetics_ 41, 29–37 (1995). Article CAS Google Scholar * Nizetic, D., Figueroa, F., Dembic, Z.,
Nevo, E. & Klein, J. Major histocompatibility complex gene organization in the mole rat _Spalax ehrenbergi_: evidence for transfer of function between class II genes. _Proc. Natl Acad.
Sci. USA_ 84, 5828–5832 (1987). Article CAS Google Scholar * Vilches, C. & Parham, P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. _Annu. Rev. Immunol._
20, 217–251 (2002). Article CAS Google Scholar * Petersen, J. et al. T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. _Nat. Struct. Mol. Biol._ 21,
480–488 (2014). Article CAS Google Scholar * Henderson, K. N. et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. _Immunity_ 27,
23–34 (2007). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work has been funded in part by the Pathogenesis and the Viral Latency Programs of the Heinrich Pette
Institute, Leibniz Institute for Experimental Virology and the German Center for Infection Research (DZIF) through TTU 04.810. This project has been funded in part with federal funds from
the Frederick National Laboratory for Cancer Research, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the
Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported in part by
the Intramural Research Program of the National Institutes of Health, Frederick National Laboratory, Center for Cancer Research. W.F.G.-B. was supported by National Institute of General
Medical Sciences (T32GM007752) and the NIH (P01-AI104715 and F31AI116366). P.J.N. was supported by NIH U19 NS095774. A.H. was supported by the German Center for Infection Research (DZIF)
through an MD/PhD Stipend (TI 07.002) and via the Clinician Scientist Program of the Faculty of Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany. J.R was supported by
an Australian Research Council Laureate Fellowship (FL160100049) and R.B was supported by a Career Development Fellowship from the National Health and Medical Research Council of Australia
(APP1109901). We would like to thank H. Reid and K. Loh for their kind gift of HLA-DQ2 viral stocks. We would like to thank the NIH Tetramer Core Facility for all provided HLA class II
monomers. AUTHOR INFORMATION Author notes * Anaïs Chapel Present address: Unité HIV Inflammation et Persistance, Institut Pasteur, Paris, France AUTHORS AND AFFILIATIONS * Research
Department Virus Immunology, Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany Annika Niehrs, Wilfredo F. Garcia-Beltran, Angelique Hölzemer, Anaïs
Chapel, Laura Richert, Christian Körner, Glòria Martrus & Marcus Altfeld * German Center for Infection Research (DZIF), Partner Site Hamburg-Lübeck-Borstel-Riems, Hamburg, Germany Annika
Niehrs, Angelique Hölzemer & Marcus Altfeld * Ragon Institute of MGH, MIT, and Harvard, Cambridge, MA, USA Wilfredo F. Garcia-Beltran & Mary Carrington * Division of Biomedical
Informatics and Personalized Medicine, University of Colorado School of Medicine, Aurora, CO, USA Paul J. Norman * Department of Microbiology and Immunology, University of Colorado School of
Medicine, Aurora, CO, USA Paul J. Norman * Infection and Immunity Program and The Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University,
Clayton, Victoria, Australia Gabrielle M. Watson, Jamie Rossjohn & Richard Berry * Australian Research Council Centre of Excellence in Advanced Molecular Imaging, Monash University,
Clayton, Victoria, Australia Gabrielle M. Watson, Jamie Rossjohn & Richard Berry * First Department of Internal Medicine, University Medical Center Eppendorf, Hamburg, Germany Angelique
Hölzemer * Inserm Inria SISTM Bordeaux Population Health Research Center UMR 1219, Univ. Bordeaux, Bordeaux, France Laura Richert * Department of Microbiology and Biotechnology, University
of Hamburg, Hamburg, Germany Andreas Pommerening-Röser * One Lambda, Inc., Canoga Park, CA, USA Mikki Ozawa & Jar-How Lee * Institute of Infection and Immunity, Cardiff University School
of Medicine, Cardiff, UK Jamie Rossjohn * Basic Science Program, HLA Immunogenetics Section, Frederick National Laboratory for Cancer Research, Frederick, MD, USA Mary Carrington Authors *
Annika Niehrs View author publications You can also search for this author inPubMed Google Scholar * Wilfredo F. Garcia-Beltran View author publications You can also search for this author
inPubMed Google Scholar * Paul J. Norman View author publications You can also search for this author inPubMed Google Scholar * Gabrielle M. Watson View author publications You can also
search for this author inPubMed Google Scholar * Angelique Hölzemer View author publications You can also search for this author inPubMed Google Scholar * Anaïs Chapel View author
publications You can also search for this author inPubMed Google Scholar * Laura Richert View author publications You can also search for this author inPubMed Google Scholar * Andreas
Pommerening-Röser View author publications You can also search for this author inPubMed Google Scholar * Christian Körner View author publications You can also search for this author
inPubMed Google Scholar * Mikki Ozawa View author publications You can also search for this author inPubMed Google Scholar * Glòria Martrus View author publications You can also search for
this author inPubMed Google Scholar * Jamie Rossjohn View author publications You can also search for this author inPubMed Google Scholar * Jar-How Lee View author publications You can also
search for this author inPubMed Google Scholar * Richard Berry View author publications You can also search for this author inPubMed Google Scholar * Mary Carrington View author publications
You can also search for this author inPubMed Google Scholar * Marcus Altfeld View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS A.N.
performed reporter cell and primary NK cell experiments and analyzed the data. A.C. and A.N. conducted and analyzed the HLA class II-coated bead assay. A.N., W.F.G.-B. and A.H. designed and
generated the Jurkat reporter cell lines. G.M.W., R.B. and J.R. conducted the SPR measurements. P.J.N., M.O. and J.-H.L. provided the single HLA-DP antigens and gave important intellectual
input. L.R. performed mixed effects linear regression models and provided important statistical guidance. A.N., W.F.G.-B. and M.A. designed the experiments. G.M., C.K., A.P.-R. and M.C. gave
important intellectual input throughout the process. M.A. supervised the study. A.N. wrote the first draft of the manuscript and M.A. revised and edited the manuscript. All authors revised
the manuscript and approved it for publication. CORRESPONDING AUTHOR Correspondence to Marcus Altfeld. ETHICS DECLARATIONS COMPETING INTERESTS A.N., W.F.G.-B. and M.A. filed a patent
application (EP18174760.1) regarding the therapeutic use of anti-NKp44 antibodies for the treatment and/or prevention of graft-versus-host disease. M.O. and J.-H.L. are current employees of
OneLambda Inc., a part of Thermo Fisher Scientific. All other authors declare no competing interest. ADDITIONAL INFORMATION PEER REVIEW INFORMATION: Zoltan Fehervari was the primary editor
on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. PUBLISHER’S NOTE: Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. INTEGRATED SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 KIR3DL1 AND KIR2DL4 FC CONSTRUCTS EXHIBIT UNSPECIFIC
BINDING TO HLA CLASS II COATED BEADS AT HIGH CONCENTRATIONS. A Binding of KIR3DL1 Fc construct at various concentrations (1–100 µg/mL) to HLA-DR (blue), HLA-DQ (yellow) and HLA-DP (red)
coated beads as well as positive (grey) and negative (black) control beads is plotted as median fluorescence intensity (MFI). Each dot represents the measured MFI for binding of the KIR3DL1
Fc construct at a specific concentration to a certain HLA class II coated bead or negative/positive control beads. Lines connect matching MFI values for one specific HLA class II allotype or
negative/positive control beads measured at different KIR3DL1 Fc construct concentrations. Data is representative for one single experiment (n = 1). B MFI values of KIR2DL4 Fc construct
binding at varying concentrations (1–100 µg/mL; left panel) and at 10 µg/mL to HLA-DR (blue), HLA-DQ (yellow) and HLA-DP (red) coated beads as well as positive (grey) and negative (black)
control beads (right panel) are depicted. Each dot in both panels represents one HLA class II molecule. Lines in the left panel connect matching MFI values for one specific HLA class II
allotype or negative/positive control beads measured at different KIR2DL4 Fc construct concentrations. Horizontal line in right panel indicates median, error bars indicate interquartile
range. Data is representative for one single experiment (n = 1). SUPPLEMENTARY FIGURE 2 UNTRANSDUCED JURKAT CELLS DO NOT SHOW FUNCTIONAL RESPONSES TO HLA CLASS II MOLECULES. Activation of
untransduced Jurkat cells in response to anti-KIR2DL3, anti-NKp46, anti-NKp44 as well as HLA-DR7 and HLA-DP401 CLIP monomers was assessed by the expression of CD69. Plot represents one of
eight independent experiments (left panel). The percentage of CD69+ cells following incubation on non-coated wells (blank) was subtracted from all samples. Corrected values are illustrated
as median with interquartile range as determined in eight independent biological replicates (n = 8). SUPPLEMENTARY FIGURE 3 UNSTIMULATED NK CELLS DO NOT EXPRESS NKP44 AND DO NOT DEGRANULATE
UPON CO-INCUBATION WITH HLA-DP401. A Surface expression of NKp44 was determined in freshly isolated untreated (black) and IL-2 plus IL-15 treated (red) primary NK cells. Plot represents one
of seven individual donors. MFI of NKp44 expression from untreated and cytokine-treated primary NK cells was determined in seven individual donors (n = 7). Each dot represents one donor.
Horizontal line indicates the median, error bars display interquartile range. Two-tailed Wilcoxon matched-pairs signed rank test was used to assess differences in the surface expression of
NKp44 between untreated and cytokine-treated primary NK cells. _*p_ = _0.02_. B Freshly isolated unstimulated NK cells were isolated from seven individual donors and co-incubated with
plate-coated anti-NKp44, HLA-DR7 CLIP, HLA-DP401 CLIP or non-coated wells (blank) in the presence of purified mouse IgG1 isotype or purified anti-human NKp44 antibody (both at a final
concentration of 10 µg/mL). The percentage of CD107a+ cells was determined. Each dot represents one individual donors (n = 7) and lines connect responses from one individual donor.
SUPPLEMENTARY FIGURE 4 HLA-DP SURFACE EXPRESSION OF JE6.1-DP TRANSDUCED CELL LINES IS INCREASED BY CLIP PULSING. A HLA-DP surface expression of HLA-DP transduced cell lines (red and blue
histograms) is depicted. The HLA-DP expression of the untransduced parental cell line is displayed in grey. Plots represent one of seven individual experiments. B HLA-DP (left panel) and
CLIP (right panel) surface expression following CLIP pulsing in comparison to DMSO-treated cells is depicted as fold change in MFI [MFI CLIP pulsed/MFI DMSO] for the four indicated JE6.1-DP
expressing cell lines. Each dot represents one individual biological replicate as determined in seven independent experiments (n = 7). Boxes represent 25th to 75th percentiles. Whiskers
indicate minimum and maximum values, horizontal line indicates the median. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figs. 1–4 and Supplementary Tables 1 and 2
REPORTING SUMMARY RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Niehrs, A., Garcia-Beltran, W.F., Norman, P.J. _et al._ A subset of HLA-DP molecules
serve as ligands for the natural cytotoxicity receptor NKp44. _Nat Immunol_ 20, 1129–1137 (2019). https://doi.org/10.1038/s41590-019-0448-4 Download citation * Received: 13 September 2018 *
Accepted: 06 June 2019 * Published: 29 July 2019 * Issue Date: September 2019 * DOI: https://doi.org/10.1038/s41590-019-0448-4 SHARE THIS ARTICLE Anyone you share the following link with
will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative
