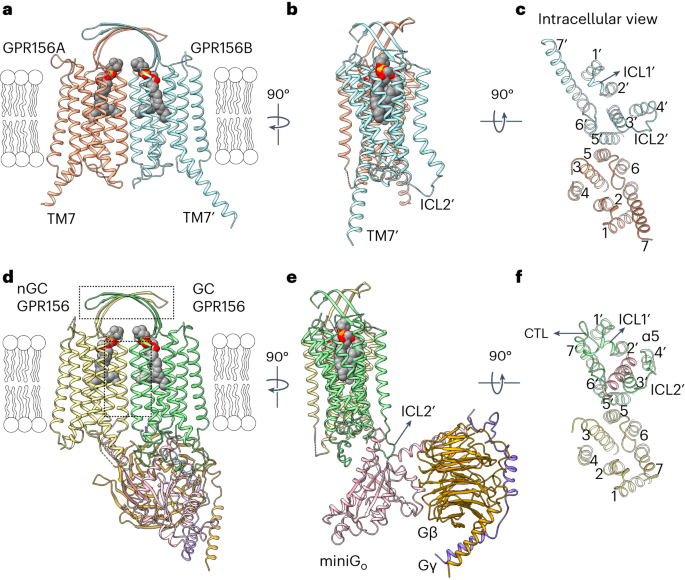
Constitutive activation mechanism of a class c gpcr
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Class C G-protein-coupled receptors (GPCRs) are activated through binding of agonists to the large extracellular domain (ECD) followed by rearrangement of the transmembrane domains
(TMDs). GPR156, a class C orphan GPCR, is unique because it lacks an ECD and exhibits constitutive activity. Impaired GPR156–Gi signaling contributes to loss of hearing. Here we present the
cryo-electron microscopy structures of human GPR156 in the Go-free and Go-coupled states. We found that an endogenous phospholipid molecule is located within each TMD of the GPR156 dimer.
Asymmetric binding of Gα to the phospholipid-bound GPR156 dimer restructures the first and second intracellular loops and the carboxy-terminal part of the elongated transmembrane 7 (TM7)
without altering dimer conformation. Our findings reveal that GPR156 is a transducer for phospholipid signaling. Constant binding of abundant phospholipid molecules and the G-protein-induced
reshaping of the cytoplasmic face provide a basis for the constitutive activation of GPR156. Access through your institution Buy or subscribe This is a preview of subscription content,
access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99
/ 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS MOLECULAR INSIGHTS INTO THE ACTIVATION MECHANISM OF GPR156 IN MAINTAINING
AUDITORY FUNCTION Article Open access 05 December 2024 SPECIFIC BINDING OF GPR174 BY ENDOGENOUS LYSOPHOSPHATIDYLSERINE LEADS TO HIGH CONSTITUTIVE GS SIGNALING Article Open access 22
September 2023 GPR161 STRUCTURE UNCOVERS THE REDUNDANT ROLE OF STEROL-REGULATED CILIARY CAMP SIGNALING IN THE HEDGEHOG PATHWAY Article 07 February 2024 DATA AVAILABILITY Atomic coordinates
and the cryo-EM map have been deposited in the the EM Data Bank (EMD) and Protein Data Bank (PDB), respectively, under the following accession numbers: EMD-35380 and PDB 8IED
(GPR156-Go–scFv16), EMD-35377 and PDB 8IEB (GPR156 dimer of GPR156-Go–scFv16), EMD-35378 and PDB 8IEC (Go–scFv16 of GPR156-Go–scFv16), EMD-35390 and PDB 8IEQ (GPR156A/B/C/D), EMD-35382 and
PDB 8IEI (GPR156A/B of GPR156A/B/C/D) and EMD-35389 and PDB 8IEP (GPR156C/D of GPR156A/B/C/D). Mass spectroscopy data are deposited on Figshare (https://doi.org/10.6084/m9.figshare.24212226
and https://doi.org/10.6084/m9.figshare.24715704.v1). The trajectories for GPR156-PC and GPR156-PG from the molecular dynamics simulations data are deposited on Zenodo
(https://doi.org/10.5281/zenodo.8418994 and https://doi.org/10.5281/zenodo.8419006, respectively). Previously published PDBs used in this study are available under PDB accession codes 7UM5,
7MTR, 7MTS, 6UO8, 7EB2, 7C7S, 7E9H, 7EPA, 7EWL, 7M3J, 7M3F, 6WIV and 7E9G. The AlphaFold2 model is available in ModelArchive (https://www.modelarchive.org) with accession code ma-1015e.
Sequence data used in the alignment for Extended Data Fig. 7 are _H. sapiens_ GPR156, GABR1, GABR2, CaSR, mGlu1, mGlu2, mGlu3, mGlu4, mGlu5 and mGlu7 (Uniprot accession codes Q8NFN8, Q9UBS5,
O75899, P41180, Q13255, Q14416, Q14832, Q14833, P41594 and Q14831, respectively). Sequence data used in the alignment for Extended Data Fig. 10d are _H. sapiens_ Gi1, Gi2, Gi3, Gs, Gq, G12
and G13 (Uniprot accession codes P63096, P04899, P08754, P63092, P50148, Q03113 and Q14344, respectively). Source data are provided with this paper. REFERENCES * Gilman, A. G. G proteins:
transducers of receptor-generated signals. _Annu. Rev. Biochem._ 56, 615–649 (1987). CAS PubMed Google Scholar * Weis, W. I. & Kobilka, B. K. The molecular basis of G protein-coupled
receptor activation. _Annu. Rev. Biochem._ 87, 897–919 (2018). CAS PubMed PubMed Central Google Scholar * Chun, L., Zhang, W. & Liu, J. Structure and ligand recognition of class C
GPCRs. _Acta Pharmacol. Sin._ 33, 312–323 (2012). CAS PubMed PubMed Central Google Scholar * Ellaithy, A., Gonzalez-Maeso, J., Logothetis, D. A. & Levitz, J. Structural and
biophysical mechanisms of class C G protein-coupled receptor function. _Trends Biochem. Sci._ 45, 1049–1064 (2020). CAS PubMed PubMed Central Google Scholar * Shen, C. et al. Structural
basis of GABAB receptor–Gi protein coupling. _Nature_ 594, 594–598 (2021). CAS PubMed PubMed Central Google Scholar * Seven, A. B. et al. G-protein activation by a metabotropic glutamate
receptor. _Nature_ 595, 450–454 (2021). CAS PubMed PubMed Central Google Scholar * Lin, S. et al. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. _Nature_ 594,
583–588 (2021). CAS PubMed Google Scholar * Watkins, L. R. & Orlandi, C. In vitro profiling of orphan G protein coupled receptor (GPCR) constitutive activity. _Br. J. Pharmacol._ 178,
2963–2975 (2021). CAS PubMed Google Scholar * Tsutsumi, N. et al. Structural basis for the constitutive activity and immunomodulatory properties of the Epstein–Barr virus-encoded G
protein-coupled receptor BILF1. _Immunity_ 54, 1405–1416.e7 (2021). CAS PubMed PubMed Central Google Scholar * Kang, Y. et al. Cryo-EM structure of human rhodopsin bound to an inhibitory
G protein. _Nature_ 558, 553–558 (2018). CAS PubMed PubMed Central Google Scholar * Lin, X. et al. Cryo-EM structures of orphan GPR21 signaling complexes. _Nat. Commun._ 14, 216 (2023).
CAS PubMed PubMed Central Google Scholar * Xu, L. et al. Cryo-EM structure of constitutively active human Frizzled 7 in complex with heterotrimeric Gs. _Cell Res._ 31, 1311–1314 (2021).
CAS PubMed PubMed Central Google Scholar * Zhang, H. et al. Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5. _Nat. Commun._ 12, 4151
(2021). CAS PubMed PubMed Central Google Scholar * Lin, X. et al. Structural basis of ligand recognition and self-activation of orphan GPR52. _Nature_ 579, 152–157 (2020). CAS PubMed
Google Scholar * Xu, P. et al. Structural insights into the lipid and ligand regulation of serotonin receptors. _Nature_ 592, 469–473 (2021). CAS PubMed Google Scholar * Xu, P. et al.
Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119. _Nat. Struct. Mol. Biol._ 29, 863–870 (2022). CAS PubMed Google Scholar * Qu, X. et
al. Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. _Nature_ 604, 779–785 (2022). CAS PubMed PubMed Central Google Scholar * Ping, Y.-Q. et al. Structural
basis for the tethered peptide activation of adhesion GPCRs. _Nature_ 604, 763–770 (2022). CAS PubMed Google Scholar * Xiao, P. et al. Tethered peptide activation mechanism of the
adhesion GPCRs ADGRG2 and ADGRG4. _Nature_ 604, 771–777 (2022). CAS PubMed Google Scholar * Barros-Álvarez, X. et al. The tethered peptide activation mechanism of adhesion GPCRs. _Nature_
604, 757–762 (2022). PubMed PubMed Central Google Scholar * Jeong, E., Kim, Y., Jeong, J. & Cho, Y. Structure of the class C orphan GPCR GPR158 in complex with RGS7-Gβ5. _Nat.
Commun._ 12, 6805 (2021). CAS PubMed PubMed Central Google Scholar * Smith, E. L. et al. GPRC5D is a target for the immunotherapy of multiple myeloma with rationally designed CAR T
cells. _Sci. Transl. Med._ 11, eaau7746 (2019). PubMed PubMed Central Google Scholar * Patil, D. N. et al. Cryo-EM structure of human GPR158 receptor coupled to the RGS7-Gβ5 signaling
complex. _Science_ 375, 86–91 (2021). PubMed PubMed Central Google Scholar * Calver, A. R. et al. Molecular cloning and characterisation of a novel GABAB-related G-protein coupled
receptor. _Mol. Brain. Res._ 110, 305–317 (2003). CAS PubMed Google Scholar * Kindt, K. S. et al. EMX2-GPR156-Gαi reverses hair cell orientation in mechanosensory epithelia. _Nat.
Commun._ 12, 2861 (2021). CAS PubMed PubMed Central Google Scholar * Greene, D. et al. Genetic association analysis of 77,539 genomes reveals rare disease etiologies. _Nat. Med._ 29,
679–688 (2023). CAS PubMed PubMed Central Google Scholar * Ramzan, M. et al. Novel GPR156 variants confirm its role in moderate sensorineural hearing loss. _Sci. Rep._ 13, 17010 (2023).
CAS PubMed PubMed Central Google Scholar * Kalam, H. et al. Identification of host regulators of _Mycobacterium tuberculosis_ phenotypes uncovers a role for the MMGT1-GPR156 lipid
droplet axis in persistence. _Cell Host Microbe_ 31, 978–992 (2023). CAS PubMed Google Scholar * Maeda, S. et al. Development of an antibody fragment that stabilizes GPCR/G-protein
complexes. _Nat. Commun._ 9, 3712 (2018). PubMed PubMed Central Google Scholar * Park, J. et al. Structure of human GABAB receptor in an inactive state. _Nature_ 584, 304–309 (2020). CAS
PubMed PubMed Central Google Scholar * Papasergi-Scott, M. M. et al. Structures of metabotropic GABAB receptor. _Nature_ 584, 310–314 (2020). CAS PubMed PubMed Central Google Scholar
* Shaye, H. et al. Structural basis of the activation of a metabotropic GABA receptor. _Nature_ 584, 298–303 (2020). CAS PubMed PubMed Central Google Scholar * Mao, C. et al. Cryo-EM
structures of inactive and active GABAB receptor. _Cell Res._ 30, 564–573 (2020). CAS PubMed PubMed Central Google Scholar * Kim, Y., Jeong, E., Jeong, J.-H., Kim, Y. & Cho, Y.
Structural basis for activation of the heterodimeric GABAB Receptor. _J. Mol. Biol._ 432, 5966–5984 (2020). CAS PubMed Google Scholar * Koehl, A. et al. Structural insights into the
activation of metabotropic glutamate receptors. _Nature_ 566, 79–84 (2019). CAS PubMed PubMed Central Google Scholar * Gao, Y. et al. Asymmetric activation of the calcium-sensing
receptor homodimer. _Nature_ 595, 455–459 (2021). CAS PubMed PubMed Central Google Scholar * Du, J. et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers. _Nature_ 594,
589–593 (2021). CAS PubMed Google Scholar * Jurcik, A. et al. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics
trajectories. _Bioinformatics_ 34, 3586–3588 (2018). CAS PubMed PubMed Central Google Scholar * Suckau, D. et al. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. _Anal.
Bioanal. Chem._ 376, 952–965 (2003). CAS PubMed Google Scholar * Carlson, M. L. et al. The peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. _eLife_
7, e34085 (2018). PubMed PubMed Central Google Scholar * Symons, J. L. et al. Lipidomic atlas of mammalian cell membranes reveals hierarchical variation induced by culture conditions,
subcellular membranes, and cell lineages. _Soft Matter_ 17, 288–297 (2021). CAS PubMed PubMed Central Google Scholar * Pin, J.-P., Galvez, T. & Prézeau, L. Evolution, structure, and
activation mechanism of family 3/C G-protein-coupled receptors. _Pharmacol. Ther._ 98, 325–354 (2003). CAS PubMed Google Scholar * Congreve, M., Oswald, C. & Marshall, F. H. Applying
structure-based drug design approaches to allosteric modulators of GPCRs. _Trends Pharmacol. Sci._ 38, 837–847 (2017). CAS PubMed Google Scholar * Potterton, L. et al. CCP4i2: the new
graphical user interface to the CCP4 program suite. _Acta Crystallogr. D. Struct. Biol._ 74, 68–84 (2018). CAS PubMed PubMed Central Google Scholar * Jumper, J. et al. Highly accurate
protein structure prediction with AlphaFold. _Nature_ 596, 583–589 (2021). CAS PubMed PubMed Central Google Scholar * Pándy-Szekeres, G. et al. GPCRdb in 2023: state-specific structure
models using AlphaFold2 and new ligand resources. _Nucleic Acids Res._ 51, D395–D402 (2022). PubMed Central Google Scholar * Flock, T. et al. Universal allosteric mechanism for Gα
activation by GPCRs. _Nature_ 524, 173–179 (2015). CAS PubMed PubMed Central Google Scholar * Nehmé, R. et al. Mini-G proteins: novel tools for studying GPCRs in their active
conformation. _PLoS One_ 12, e0175642 (2017). PubMed PubMed Central Google Scholar * Liang, J. et al. Structural basis of lysophosphatidylserine receptor GPR174 ligand recognition and
activation. _Nat. Commun._ 14, 1012 (2023). CAS PubMed PubMed Central Google Scholar * Koehl, A. et al. Structure of the µ-opioid receptor–Gi protein complex. _Nature_ 558, 547–552
(2018). CAS PubMed PubMed Central Google Scholar * Kim, K. et al. Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. _Cell_ 182, 1574–1588.e19 (2020). CAS
PubMed PubMed Central Google Scholar * Zhang, S. et al. Inactive and active state structures template selective tools for the human 5-HT5A receptor. _Nat. Struct. Mol. Biol._ 29, 677–687
(2022). CAS PubMed PubMed Central Google Scholar * Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure
determination. _Nat. Methods_ 14, 290–296 (2017). CAS PubMed Google Scholar * Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. _J.
Struct. Biol._ 192, 216–221 (2015). PubMed PubMed Central Google Scholar * Yue, Y. et al. Structural insight into apelin receptor-G protein stoichiometry. _Nat. Struct. Mol. Biol._ 29,
688–697 (2022). CAS PubMed Google Scholar * Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. _Nat. Methods_ 16,
1153–1160 (2018). Google Scholar * Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. _Commun. Biol._ 4, 874 (2021). PubMed PubMed Central
Google Scholar * Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. _J. Comput. Chem._ 25, 1605–1612 (2004). CAS PubMed Google Scholar *
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. _Acta Crystallogr. D. Biol. Crystallogr._ 60, 2126–2132 (2004). PubMed Google Scholar * Afonine, P. V. et
al. Real-space refinement in PHENIX for cryo-EM and crystallography. _Acta Crystallogr. D. Struct. Biol._ 74, 531–544 (2018). CAS PubMed PubMed Central Google Scholar * Adams, P. D. et
al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. _Acta Crystallogr. D. Biol. Crystallogr._ 66, 213–221 (2010). CAS PubMed PubMed Central Google
Scholar * Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. _Acta Crystallogr. D. Biol. Crystallogr._ 66, 12–21 (2010). CAS PubMed Google
Scholar * Moriarty, N. W., Grosse‐Kunstleve, R. W. & Adams, P. D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation.
_Acta Crystallogr. D. Biol. Crystallogr._ 65, 1074–1080 (2009). CAS PubMed PubMed Central Google Scholar * Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps
and atomic models. _Acta Crystallogr. D. Struct. Biol._ 74, 814–840 (2018). CAS PubMed PubMed Central Google Scholar * Olsen, R. H. J. et al. TRUPATH, an open-source biosensor platform
for interrogating the GPCR transducerome. _Nat. Chem. Biol._ 16, 841–849 (2020). CAS PubMed PubMed Central Google Scholar * Schmidpeter, P. A. M. et al. Anionic lipids unlock the gates
of select ion channels in the pacemaker family. _Nat. Struct. Mol. Biol._ 29, 1092–1100 (2022). CAS PubMed PubMed Central Google Scholar * Hutchins, P. D., Russell, J. D. & Coon, J.
J. LipiDex: an integrated software package for high-confidence lipid identification. _Cell Syst._ 6, 621–625.e5 (2018). CAS PubMed PubMed Central Google Scholar * Pluskal, T., Castillo,
S., Villar-Briones, A. & Orešič, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. _BMC Bioinform._ 11, 395
(2010). Google Scholar * An, J. N. et al. Effects of periostin deficiency on kidney aging and lipid metabolism. _Aging (Albany NY)_ 13, 22649–22665 (2021). CAS PubMed Google Scholar *
Breil, C., Vian, M. A., Zemb, T., Kunz, W. & Chemat, F. ‘Bligh and Dyer’ and Folch methods for solid–liquid–liquid extraction of lipids from microorganisms. Comprehension of solvatation
mechanisms and towards substitution with alternative solvents. _Int. J. Mol. Sci._ 18, 708 (2017). PubMed PubMed Central Google Scholar * Lee, J. W., Nishiumi, S., Yoshida, M., Fukusaki,
E. & Bamba, T. Simultaneous profiling of polar lipids by supercritical fluid chromatography/tandem mass spectrometry with methylation. _J. Chromatogr. A_ 1279, 98–107 (2013). CAS PubMed
Google Scholar * Lee, J. W. et al. Detailed characterization of alterations in the lipid profiles during autophagic cell death of leukemia cells. _RSC Adv._ 6, 29512–29518 (2016). CAS
Google Scholar * Shanta, S. R. et al. Binary matrix for MALDI imaging mass spectrometry of phospholipids in both ion modes. _Anal. Chem._ 83, 1252–1259 (2011). CAS PubMed Google Scholar
* Fahy, E., Sud, M., Cotter, D. & Subramaniam, S. LIPID MAPS online tools for lipid research. _Nucleic Acids Res._ 35, W606–W612 (2007). PubMed PubMed Central Google Scholar * Sud, M.
et al. LMSD: LIPID MAPS structure database. _Nucleic Acids Res._ 35, D527–D532 (2007). CAS PubMed Google Scholar * Noh, S. A. et al. Alterations in lipid profile of the aging kidney
identified by MALDI imaging mass spectrometry. _J. Proteome Res._ 18, 2803–2812 (2019). CAS PubMed Google Scholar * Liebisch, G. et al. Quantitative measurement of different ceramide
species from crude cellular extracts by electrospray ionization tandem mass spectrometry (ESI–MS/MS). _J. Lipid Res._ 40, 1539–1546 (1999). CAS PubMed Google Scholar * Hsu, F.-F. &
Turk, J. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. _J. Am. Soc. Mass Spectr._ 11, 437–449 (2000). CAS Google Scholar * Pi, J., Wu,
X. & Feng, Y. Fragmentation patterns of five types of phospholipids by ultra-high-performance liquid chromatography electrospray ionization quadrupole time-of-flight tandem mass
spectrometry. _Anal. Methods_ 8, 1319–1332 (2016). CAS Google Scholar * Sugawara, T., Aida, K., Duan, J. & Hirata, T. Analysis of glucosylceramides from various sources by liquid
chromatography–ion trap mass spectrometry. _J. Oleo Sci._ 59, 387–394 (2010). CAS PubMed Google Scholar * Gu, M., Kerwin, J. L., Watts, J. D. & Aebersold, R. Ceramide profiling of
complex lipid mixtures by electrospray ionization mass spectrometry. _Anal. Biochem._ 244, 347–356 (1997). CAS PubMed Google Scholar * Abraham, M. J. et al. GROMACS: high performance
molecular simulations through multi-level parallelism from laptops to supercomputers. _SoftwareX_ 1, 19–25 (2015). Google Scholar * Huang, J. et al. CHARMM36m: an improved force field for
folded and intrinsically disordered proteins. _Nat. Methods_ 14, 71–73 (2017). CAS PubMed Google Scholar * Abagyan, R., Totrov, M. & Kuznetsov, D. ICM—a new method for protein
modeling and design: applications to docking and structure prediction from the distorted native conformation. _J. Comput. Chem._ 15, 488–506 (1994). CAS Google Scholar * Jo, S., Kim, T.,
Iyer, V. G. & Im, W. CHARMM‐GUI: a web‐based graphical user interface for CHARMM. _J. Comput. Chem._ 29, 1859–1865 (2008). CAS PubMed Google Scholar * Lomize, M. A., Pogozheva, I. D.,
Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. _Nucleic Acids Res._ 40, D370–D376 (2012). CAS PubMed
Google Scholar * Wu, E. L. et al. CHARMM‐GUI Membrane Builder toward realistic biological membrane simulations. _J. Comput. Chem._ 35, 1997–2004 (2014). CAS PubMed PubMed Central Google
Scholar * Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. _J. Comput. Chem._ 18, 1463–1472 (1997). CAS
Google Scholar * Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. _J. Chem. Phys._ 126, 014101 (2007). PubMed Google Scholar * McGibbon, R. T. et
al. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. _Biophys. J._ 109, 1528–1532 (2015). CAS PubMed PubMed Central Google Scholar * Michaud‐Agrawal,
N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. _J. Comput. Chem._ 32, 2319–2327 (2011). PubMed PubMed Central
Google Scholar * Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. _Nat. Methods_ 17, 261–272 (2020). CAS PubMed PubMed Central Google Scholar
Download references ACKNOWLEDGEMENTS We thank J. Koh for technical help and Y. Kim, C. Lee, J. Lee, K. Kim, S. H. Ryu (POSTECH), Y. Yu (Kookmin U.), M. Jin (GIST) and J. Kim (SNU) for
helpful comments. This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean government (MEST, No. 2021R1A2C301335711 and 2019M3E5D6066058 to
Y.C.), the Bio & Medical Technology Development Program (NRF-2019M3E5D3073567 to K.P.K.), the Ministry of Science and ICT (grant number 2022R1A2C1005885 to J.H.), the BK21 program
(Ministry of Education to Y.C.), Wellcome Trust Investigator Award (221795/Z/20/Z to X.Q., D.W., C.V.R.) and internal funding from the University of Southern California Dornsife College (to
V.K). Cryo-EM data were acquired at the Core Research Facility, Pusan National University. Computing resources were provided by the Center for Advanced Research Computing (CARC) at the
University of Southern California (https://carc.usc.edu). AUTHOR INFORMATION Author notes * These authors contributed equally: Jinwoo Shin, Junhyeon Park. AUTHORS AND AFFILIATIONS *
Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea Jinwoo Shin, Junhyeon Park & Yunje Cho * Department of Applied Chemistry, Global
Center for Pharmaceutical Ingredient Materials, Kyung Hee University, Yongin, Republic of Korea Jieun Jeong & Kwang Pyo Kim * Department of Quantitative and Computational Biology,
University of Southern California, Los Angeles, CA, USA Jordy Homing Lam & Vsevolod Katritch * Bridge Institute and Michelson Center for Convergent Biosciences, University of Southern
California, Los Angeles, CA, USA Jordy Homing Lam & Vsevolod Katritch * Department of Chemistry, University of Oxford, Oxford, UK Xingyu Qiu, Di Wu & Carol V. Robinson * Kavli
Institute for Nanoscience Discovery, University of Oxford, Oxford, UK Xingyu Qiu, Di Wu & Carol V. Robinson * Department of Pharmacy, Yonsei University, Incheon, Republic of Korea Kuglae
Kim * Therapeutics and Biotechnology Division, Korea Research Institute of Chemical Technology, 141 Gajeong-ro, Yuseong-gu, Daejeon, Republic of Korea Joo-Youn Lee * School of Pharmacy,
Sungkyunkwan University, Suwon, Republic of Korea Jaekyung Hyun * Center for New Technologies in Drug Discovery and Development, University of Southern California, Los Angeles, CA, USA
Vsevolod Katritch * Department of Chemistry, University of Southern California, Los Angeles, CA, USA Vsevolod Katritch * Department of Biomedical Science and Technology, Kyung Hee Medical
Science Research Institute, Kyung Hee University, Seoul, Republic of Korea Kwang Pyo Kim * Department of Medical Science and Engineering, Pohang University of Science and Technology, Pohang,
Republic of Korea Yunje Cho Authors * Jinwoo Shin View author publications You can also search for this author inPubMed Google Scholar * Junhyeon Park View author publications You can also
search for this author inPubMed Google Scholar * Jieun Jeong View author publications You can also search for this author inPubMed Google Scholar * Jordy Homing Lam View author publications
You can also search for this author inPubMed Google Scholar * Xingyu Qiu View author publications You can also search for this author inPubMed Google Scholar * Di Wu View author publications
You can also search for this author inPubMed Google Scholar * Kuglae Kim View author publications You can also search for this author inPubMed Google Scholar * Joo-Youn Lee View author
publications You can also search for this author inPubMed Google Scholar * Carol V. Robinson View author publications You can also search for this author inPubMed Google Scholar * Jaekyung
Hyun View author publications You can also search for this author inPubMed Google Scholar * Vsevolod Katritch View author publications You can also search for this author inPubMed Google
Scholar * Kwang Pyo Kim View author publications You can also search for this author inPubMed Google Scholar * Yunje Cho View author publications You can also search for this author inPubMed
Google Scholar CONTRIBUTIONS J.S. carried out protein expression, purification and structure determination with the help of J.P. J.S. carried out data collection with the help of J.H. J.P.
and J.S. performed biochemical experiments with the help of K.K. and J.-Y.L. K.P.K. and J.J. performed mass spectroscopy and phospholipid characterization analysis. X.Q., D.W. and C.V.R.
performed comparative lipidomics analysis. J.H.L. and V.K. performed molecular dynamics simulations. J.S., J.P. and Y.C. designed the research; Y.C. wrote the manuscript with the help of
J.S., J.P. and K.P.K. CORRESPONDING AUTHORS Correspondence to Kwang Pyo Kim or Yunje Cho. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER
REVIEW INFORMATION _Nature Structural & Molecular Biology_ thanks Bryan Roth and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer
reports are available. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the _Nature Structural & Molecular Biology_ team. ADDITIONAL INFORMATION PUBLISHER’S NOTE
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 PURIFICATION OF THE GPR156-GO
COMPLEX. A–C, Size-exclusion chromatography profile (A), SDS-PAGE gel (B), and negative-staining microscopy analysis (C) of the purified GPR156-Go complex. Purification experiments of the
GPR156-Go complex in (B) were performed independently at least three times. Negative-staining microscopy analysis (C) was performed once. D, Representative cryo-EM micrograph of the
GPR156-Go complex. The cryo-EM data collection of GPR156-Go was performed once. E, 2D class averages of the Go-free GPR156. F, 2D class averages of the GPR156-Go complex. G, Comparison of
constitutive activities of GPR156 and GABAB in G-protein activation, measured by BRET2 assay. The activity of GABAB was observed in the absence or presence of 100 µM GABA. Data were mean ±
SEM from three independent experiments, performed in technical triplicate. Statistical differences in ΔBRET were analyzed by two-way ANOVA with Dunnett’s post hoc test. H, cAMP inhibition
assay for GPR156 C-terminal truncation. Mock transfected with an empty vector was used as a negative control. cAMP production was normalized to a percent of WT activity. I, Surface
expression levels of WT and GPR156 mutants in the cAMP assay, measured by ELISA. Surface expression levels of GPR156 mutants were normalized to a percent of WT surface expression level. Data
in H and I were mean ± SEM from at least three independent experiments, performed in technical triplicate. Statistical differences were analyzed by one-way ANOVA with Dunnett’s post hoc
test, compared to WT (NS, not significant; ***_P_ < 0.001; ****_P_ < 0.0001). J, K, A model of GPR156 predicted from AlphaFold2 was used for initial docking and model building. The
N-terminal (1 to 39) and the C-terminal (336 to 814) regions are disordered and omitted in the figure. The model is colored according to the predicted Local Distance Difference Test (pLDDT)
score (J). The Predicted-Alignment-Error (PAE) plot of the GPR156 model (K). Source data EXTENDED DATA FIG. 2 FLOW CHART OF CRYO-EM AND DATA PROCESSING. Cryo-EM processing chart of the
G-protein coupled and G-protein free GPR156. EXTENDED DATA FIG. 3 ANALYSIS OF THE QUALITY OF THE CRYO-EM MAP. A, B, Angular distributions, Fourier shell correlation curves, and globally
refined cryo-EM density maps of GPR156-Go (A) and GPR156 alone (B). C–F, Angular distributions, Fourier shell correlation curves and locally refined cryo-EM density maps marked local
resolution of GPR156 dimer (C), G-protein-scFv16 (D), GPR156A/B (E), and GPR156C/D (F). G, H, Fourier shell correlation curves of the model versus the map generated through PHENIX.Mtriage64
of globally and locally refined GPR156-Go (G) and GPR156 alone (H). I, J, Global fitting of the structures of GPR156-Go into the composite map (I) and GPR156 alone into the globally refined
map (J). K, Cryo-EM densities and fitted atomic models. GF-GPR156, Go, GPR156A, GPR156B, and GC-GPR156 are shown in yellow, pink, salmon, cyan, and green, respectively. EXTENDED DATA FIG. 4
KEY FEATURES OF GPR156. A, Cryo-EM map of the GPR156 tetramer. B, Interface between the head-to-head dimer of GPR156. C, Interaction between TM7 of GPR156C and ICL2 of GPR156B. H-bonds
(Q314-D155 and E321-V158) and hydrophobic interactions (F318-V158 and I325-I159) are highlighted. D, E, Aligned structures of the two GPR156 dimers in two views; front view (D), bottom view.
Structures of the ICL2s are encircled (E). F–H, Structural comparison of GPR156B and GABAB-Gi (PDB:7EB2 ref. 5) with respect to ECL2 (F), TM7 (G), and ICLs (H). I, Cryo-EM map of the
GPR156-Go complex. J, K, Aligned structures of an GPR156 alone dimer with the GPR156-Go complex in two views; front view (J), bottom view (K). L, A density on top of the ECL2 in two
different views. M, Close-up view of the interactions between Y1463.55 (nGC) and H248ICL3 (GC). EXTENDED DATA FIG. 5 COMPARISON OF THE DIMERIC ARRANGEMENT OF GPR156 WITH OTHER CLASS C GPCRS
IN INACTIVE AND ACTIVE STATES. A–I, The TMDs of Class C GPCRs were aligned with GC-GPR156 (black line) and shown in the extracellular (top) view. GPR156-Go (A), inactive GABAB (red; PDB:
7C7S ref. 33) (B), active GABAB (orange; 7EB2 ref. 5) (C), active mGlu4 (beige; 7E9H ref. 7) (D), inactive mGlu2 (yellow; 7EPA ref. 37) (E), active mGlu2 (green; 7MTS ref. 6) (F), apo GPR158
(pink; 7EWL ref. 21) (G), inactive CaSR (blue; 7M3J ref. 36) (H), active CaSR (purple; 7M3F ref. 36) (I). The gray filled line represents nGC-GPR156. EXTENDED DATA FIG. 6 CHARACTERIZATION
OF PHOSPHOLIPID IN GPR156. A, Comparative lipidomics analysis of endogenous lipids bound to purified recombinant GPR156. PC and PE are enrichment in GPR156 fraction relative to total
cellular lysate. Bars show mean ± standard deviation from three independent experiments (dots). B–C, GPR156 activation as measured by GTPase-Glo assay for GPR156-Gi peptidisc containing PE
(B) or GPR156 in LMNG (C). A peptidisc containing GPRC5D-Gi and PE was incubated with PG as a control. Lower levels of residual GTP indicate higher level of G-protein activity. Data in (B)
and (C) were mean ± SEM from three independent experiments, performed in technical duplicate. Statistical differences were analyzed by one-way ANOVA with Dunnett’s post hoc test. D‒F,
Comparison of molecular dynamics simulation of GPR156 Go-coupled complexes with PG versus PC. D, Root-mean-square-fluctuation (r.m.s.f.) calculated of each subunit in the complexes; shading
refers to 95 % confidence interval (n = 5). E–F, Distribution of the closest distances between the sidechain of R2796.57 and the phospholipids against the closest distances between the
backbone of C216ECL2 and the phospholipids on the nGC protomer (E) and on the GC protomer (F). The shading refers to density estimated with a multivariate gaussian kernel; the marginal
distribution (by count) is shown on the sidebar. The black points in the background is the datapoints collected every 0.5 ns. All distances were calculated using only the heavy atoms. The
horizontal and vertical dotted line refers to 3.5 Å. G, Mutational effect on the F215ECL2 adjacent to the phospholipid head in G-protein activation, measured by BRET2 assay. Data were mean ±
SEM from four independent experiments, performed in technical triplicate. Statistical differences were analyzed by two-way ANOVA with Dunnett’s post hoc test, compared to WT (NS, not
significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). H‒I, Comparison of PC- and PG-bound GPR156 with representative snapshots from MD simulations of GPR156.
Close-up views of a phospholipid-binding site in the simulations of PC-bound (H) and PG-bound GPR156 (I). nGC protomer and GC protomer are shown on top and bottom, respectively. Three
representative snapshots (970, 985, and 1000 ns) of a simulation trajectory are displayed. Source data EXTENDED DATA FIG. 7 SEQUENCE ALIGNMENT OF GPR156 WITH OTHER HUMAN CLASS C GPCRS. The
conserved residues are marked in yellow. The alignment was output from GPCRdb (gpcrdb.org) and edited by using snapgene (snapgene.com). EXTENDED DATA FIG. 8 PHOSPHOLIPIDS IN GPR156. A, In
one protomer, the phenyl ring is flipped by 85° toward the dimer interface, creating space for the lateral movement of the phospholipid. We refer this conformer to as an open form. The W229
indole ring can be repositioned upon the conformational change of the F275 ring. B, In another protomer, F275 is packed against the fatty-acyl chain of the phospholipid to form a closed
conformation. C, Density at the top half of the dimer interface, in which a CLR molecule is modelled. D, E, Densities near V223 (D) and W284 (E) in GPR156. F, Comparison of the
phospholipid-binding in GPR156 with that of GABAB2. G, Comparison of the phospholipid-binding in GPR156 with the PAM-binding in mGlu2 and CaSR. H–J, cAMP inhibition assay for GPR156 mutated
at the phospholipid binding site (H), dimer interface (I, J). Data were mean ± SEM from at least three independent experiments, performed in technical triplicate. Statistical differences
were analyzed by one-way ANOVA with Dunnett’s post hoc test, compared to WT (NS, not significant; **P < 0.01; ***P < 0.001; ****P < 0.0001). Source data EXTENDED DATA FIG. 9
STRUCTURAL TRANSITION OF THE CYTOPLASMIC FACE OF GPR156 IN THE GO PROTEIN-COUPLED STATE. A–C, Comparison of the Go-free, nGC-, and GC-protomers in three different views; front (A),
extracellular (B), and cytoplasmic view (C). D–E, Aligned ICL1s (D) and ICL2s (E) of the GPR156B with GC-protomers. The major structural differences in the C-terminal loop and ICLs are
indicated by red arrows. F, cAMP inhibition assay for GPR156 mutated at ICL2. Data were mean ± SEM from three independent experiments. Statistical differences were analyzed by one-way ANOVA
with Dunnett’s post hoc test, compared to WT. G–L, Comparison of the TMD and ICLs of GPR156 with those of other class C GPCRs–Gi; GABAB-Gi1 (PDB: 7EB2 ref. 5), mGlu2-Gi1 (7MTS ref. 6),
mGlu4-Gi3 (7E9H ref. 7). Comparison of the TMD of GPR156 GC-protomer with those of other class C GPCRs bound to Gi in three different views; front (G), extracellular (H), and cytoplasmic
view (I). Comparison of ICL1 (J), ICL2 (K), and ICL3 and the C-terminal loop (L). M, Mutational effect on CTL of GPR156 in G-protein activation, measured by BRET2 assay. Data were mean ± SEM
from three independent experiments, performed in technical triplicate. Statistical differences were analyzed by two-way ANOVA with Dunnett’s post hoc test, compared to WT (NS, not
significant; **P < 0.01; ***P < 0.001; ****P < 0.0001). Source data EXTENDED DATA FIG. 10 GO BINDING TO GPR156. A–C, Comparison of Go binding between GPR156 and other class C GPCRs:
GABAB-Gi1 (PDB: 7EB2 ref. 5) (A), mGlu2-Gi1 (7MTS ref. 6) (B), mGlu4-Gi3 (7E9H ref. 7) (C). The red arrows indicate the structural differences in the receptors and the G proteins. D.
Sequence alignment of the residues in the α5 helix in different human Gα proteins. Residues interacting with GPR156 are marked with a light green circle. Absolutely conserved and highly
conserved (≥50%) residues are marked with orange and yellow colors, respectively. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Notes 1–7, Videos 1–3, and Tables 1–4.
REPORTING SUMMARY PEER REVIEW FILE SUPPLEMENTARY VIDEO 1 Dynamics of TM4 and ICL2 of the GPR156B protomer revealed by 3D variability analysis. SUPPLEMENTARY VIDEO 2 Flexibility of CTL in the
GPR156 GC protomer. SUPPLEMENTARY VIDEO 3 Rigid body movement of the GC protomer with respect to the G protein. SOURCE DATA SOURCE DATA FIG. 2 Statistical source data. SOURCE DATA FIG. 3
Statistical source data. SOURCE DATA FIG. 5 Statistical source data. SOURCE DATA EXTENDED DATA FIG./TABLE 1 Unprocessed gel SOURCE DATA EXTENDED DATA FIG./TABLE 1 Statistical source data.
SOURCE DATA EXTENDED DATA FIG./TABLE 6 Statistical source data. SOURCE DATA EXTENDED DATA FIG./TABLE 8 Statistical source data. SOURCE DATA EXTENDED DATA FIG./TABLE 9 Statistical source
data. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other
rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Shin, J., Park, J., Jeong, J. _et al._ Constitutive activation mechanism of a class C GPCR. _Nat Struct Mol Biol_ 31, 678–687 (2024).
https://doi.org/10.1038/s41594-024-01224-7 Download citation * Received: 22 February 2023 * Accepted: 09 January 2024 * Published: 08 February 2024 * Issue Date: April 2024 * DOI:
https://doi.org/10.1038/s41594-024-01224-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
