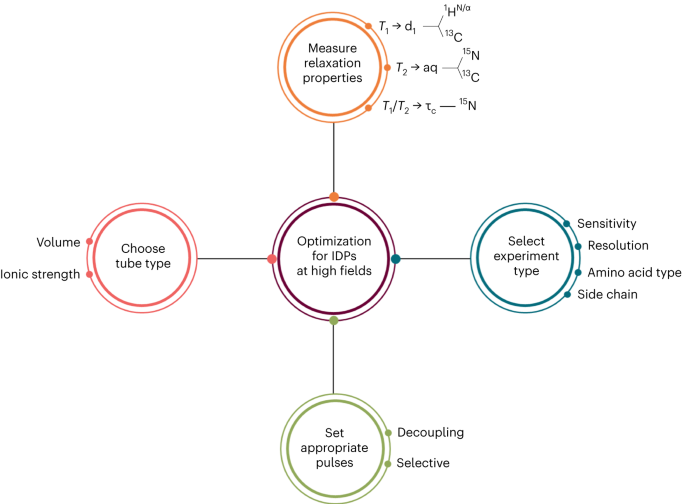
Optimal 13c nmr investigation of intrinsically disordered proteins at 1. 2 ghz
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Nuclear magnetic resonance (NMR) spectroscopy is a powerful technique for characterizing biomolecules such as proteins and nucleic acids at atomic resolution. Increased magnetic
field strengths drive progress in biomolecular NMR applications, leading to improved performance, e.g., higher resolution. A new class of NMR spectrometers with a 28.2 T magnetic field (1.2
GHz 1H frequency) has been commercially available since the end of 2019. The availability of ultra-high-field NMR instrumentation makes it possible to investigate more complex systems using
NMR. This is especially true for highly flexible intrinsically disordered proteins (IDPs) and highly flexible regions (IDRs) of complex multidomain proteins. Indeed, the investigation of
these proteins is frequently hampered by the crowding of NMR spectra. The advantages, however, are accompanied by challenges that the user must overcome when conducting experiments at such a
high field (e.g., large spectral widths, radio frequency bandwidth, performance of decoupling schemes). This protocol presents strategies and tricks for optimising high-field NMR
experiments for IDPs/IDRs based on the analysis of the relaxation properties of the investigated protein. The protocol, tested on three IDPs of different molecular weight and structural
complexity, focuses on 13C-detected NMR at 1.2 GHz. A set of experiments, including some multiple receiver experiments, and tips to implement versions tailored for IDPs/IDRs are described.
However, the general approach and most considerations can also be applied to experiments that acquire 1H or 15N nuclei and to experiments performed at lower field strengths. KEY POINTS *
28.2 T nuclear magnetic resonance spectrometers are now available and, thanks to their improved resolution, are especially useful for analyzing proteins that have flexible regions. * At such
high magnetic fields, there are important challenges relating to the concomitant increase in spectral width. Key points explored in this protocol include the relaxation properties of
proteins, choice of pulses for excitation and decoupling and setup of two-dimensional and multiple receiver experiments. Access through your institution Buy or subscribe This is a preview of
subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value
online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more
Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS:
* Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS QUANTIFYING THE THERMODYNAMICS OF PROTEIN UNFOLDING USING
2D NMR SPECTROSCOPY Article Open access 07 August 2020 CHARACTERIZING PROTEINS IN A NATIVE BACTERIAL ENVIRONMENT USING SOLID-STATE NMR SPECTROSCOPY Article 13 January 2021 ENSEMBLE
DETERMINATION BY NMR DATA DECONVOLUTION Article 11 May 2023 DATA AVAILABILITY The data are available upon request to the authors. CODE AVAILABILITY Pulse sequences are deposited at
https://doi.org/10.6084/m9.figshare.23864817. REFERENCES * Banci, L. et al. Biomolecular NMR at 1.2 GHz. Preprint at https://doi.org/10.48550/arXiv.1910.07462 (2019). * Wikus, P., Frantz,
W., Kümmerle, R. & Vonlanthen, P. Commercial gigahertz-class NMR magnets. _Supercond. Sci. Technol._ 35, 033001 (2022). Article ADS Google Scholar * Luchinat, E., Barbieri, L.,
Cremonini, M. & Banci, L. Protein in-cell NMR spectroscopy at 1.2 GHz. _J. Biomol. NMR_ 75, 97–107 (2021). Article CAS PubMed PubMed Central Google Scholar * Nimerovsky, E. et al.
Proton detected solid-state NMR of membrane proteins at 28 Tesla (1.2 GHz) and 100 KHz magic-angle spinning. _Biomolecules_ 11, 752 (2021). Article CAS PubMed PubMed Central Google
Scholar * Callon, M. et al. Biomolecular solid-state NMR spectroscopy at 1200 MHz: the gain in resolution. _J. Biomol. NMR_ 75, 255–272 (2021). Article CAS PubMed PubMed Central Google
Scholar * Schwalbe, H. et al. Structural and dynamical properties of a denatured protein. Heteronuclear 3D NMR experiments and theoretical simulations of lysozyme in 8 M urea.
_Biochemistry_ 36, 8977–8991 (1997). Article CAS PubMed Google Scholar * Mittag, T. & Forman-Kay, J. D. Atomic-level characterization of disordered protein ensembles. _Curr. Opin.
Struct. Biol._ 17, 3–14 (2007). Article CAS PubMed Google Scholar * Nováček, J., Žídek, L. & Sklenář, V. Toward optimal-resolution NMR of intrinsically disordered proteins. _J. Magn.
Reson._ 241, 41–52 (2014). Article PubMed ADS Google Scholar * Bermel, W. et al. Improving the chemical shift dispersion of multidimensional NMR spectra of intrinsically disordered
proteins. _J. Biomol. NMR_ 55, 231–237 (2013). Article CAS PubMed Google Scholar * Felli, I. C. & Pierattelli, R. Novel methods based on 13C detection to study intrinsically
disordered proteins. _J. Magn. Reson._ 241, 115–125 (2014). Article CAS PubMed ADS Google Scholar * Bermel, W., Bertini, I., Felli, I., Piccioli, M. & Pierattelli, R. 13C-detected
protonless NMR spectroscopy of proteins in solution. _Prog. Nucl. Magn. Reson. Spectrosc._ 48, 25–45 (2006). Article CAS Google Scholar * Felli, I. C. & Pierattelli, R. 13C direct
detected NMR for challenging systems. _Chem. Rev._ 122, 9468–9496 (2022). Article CAS PubMed PubMed Central Google Scholar * Bermel, W. et al. Complete assignment of heteronuclear
protein resonances by protonless NMR spectroscopy. _Angew. Chem. Int. Ed._ 44, 3089–3092 (2005). Article CAS Google Scholar * Peng, J. W. & Wagner, G. Mapping of spectral density
functions using heteronuclear NMR relaxation measurements. _J. Magn. Reson._ 98, 308–332 (1992). CAS ADS Google Scholar * Fushman, D., Tjandra, N. & Cowburn, D. An approach to direct
determination of protein dynamics from 15N NMR relaxation at multiple fields, independent of variable 15N chemical shift anisotropy and chemical exchange contributions. _J. Am. Chem. Soc._
121, 8577–8582 (1999). Article CAS Google Scholar * Lipari, G. & Szabo, A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2.
Analysis of experimental results. _J. Am. Chem. Soc._ 104, 4559–4570 (1982). Article CAS Google Scholar * Bermel, W., Bertini, I., Felli, I. C., Kümmerle, R. & Pierattelli, R. Novel
13C direct detection experiments, including extension to the third dimension, to perform the complete assignment of proteins. _J. Magn. Reson._ 178, 56–64 (2006). Article CAS PubMed ADS
Google Scholar * Bermel, W., Bertini, I., Felli, I. C. & Pierattelli, R. Speeding up 13C direct detection biomolecular NMR spectroscopy. _J. Am. Chem. Soc._ 131, 15339–15345 (2009).
Article CAS PubMed Google Scholar * Gil, S. et al. NMR spectroscopic studies of intrinsically disordered proteins at near-physiological conditions. _Angew. Chem. Int. Ed._ 52,
11808–11812 (2013). Article CAS Google Scholar * Murrali, M. G., Piai, A., Bermel, W., Felli, I. C. & Pierattelli, R. Proline fingerprint in intrinsically disordered proteins.
_Chembiochem_ 19, 1625–1629 (2018). Article CAS PubMed Google Scholar * Felli, I. C., Bermel, W. & Pierattelli, R. Exclusively heteronuclear NMR experiments for the investigation of
intrinsically disordered proteins: focusing on proline residues. _Magn. Reson._ 2, 511–522 (2021). Article CAS Google Scholar * Bermel, W. et al. H-Start for exclusively heteronuclear NMR
spectroscopy: the case of intrinsically disordered proteins. _J. Magn. Reson._ 198, 275–281 (2009). Article CAS PubMed ADS Google Scholar * Bermel, W. et al. Protonless NMR experiments
for sequence-specific assignment of backbone nuclei in unfolded proteins. _J. Am. Chem. Soc._ 128, 3918–3919 (2006). Article CAS PubMed Google Scholar * Felli, I. C. & Pierattelli,
R. Spin-state-selective methods in solution- and solid-state biomolecular 13C NMR. _Prog. Nucl. Magn. Reson. Spectrosc._ 84–85, 1–13 (2015). Article PubMed Google Scholar * Sørensen, M.
D., Meissner, A. & Sørensen, O. W. Spin-state-selective coherence transfer via intermediate states of two-spin coherence in IS spin systems: application to E.COSY-type measurement of J
coupling constants. _J. Biomol. NMR_ 10, 181–186 (1997). Article Google Scholar * Shimba, N., Stern, A. S., Craik, C. S., Hoch, J. C. & Dötsch, V. Elimination of 13Cα splitting in
protein NMR spectra by deconvolution with maximum entropy reconstruction. _J. Am. Chem. Soc._ 125, 2382–2383 (2003). Article CAS PubMed Google Scholar * Ying, J., Li, F., Lee, J. H.
& Bax, A. 13Cα decoupling during direct observation of carbonyl resonances in solution NMR of isotopically enriched proteins. _J. Biomol. NMR_ 60, 15–21 (2014). Article CAS PubMed
PubMed Central Google Scholar * Karunanithy, G., Mackenzie, H. W. & Hansen, D. F. Virtual homonuclear decoupling in direct detection nuclear magnetic resonance experiments using deep
neural networks. _J. Am. Chem. Soc._ 143, 16935–16942 (2021). Article CAS PubMed Google Scholar * Kupče, E. R. & Freeman, R. Parallel receivers and sparse sampling in
multidimensional NMR. _J. Magn. Reson._ 213, 1–13 (2011). Article PubMed ADS Google Scholar * Kupče, E. & Kay, L. E. Parallel acquisition of multi-dimensional spectra in protein NMR.
_J. Biomol. NMR_ 54, 1–7 (2012). Article PubMed Google Scholar * Kupce, E. NMR with multiple receivers. _Mod. NMR Methodol._ 4, 721–731 (2015). CAS Google Scholar * Viegas, A.,
Viennet, T., Yu, T. & Schumann, F. UTOPIA NMR: activating unexploited magnetization using interleaved low-gamma detection. _J. Biomol. NMR_ 64, 9–15 (2016). Article CAS PubMed PubMed
Central Google Scholar * Schiavina, M. et al. Taking simultaneous snapshots of intrinsically disordered proteins in action. _Biophys. J._ 117, 46–55 (2019). Article CAS PubMed PubMed
Central ADS Google Scholar * Mori, S., Abeygunawardana, C., Johnson, M. O. & Vanzijl, P. C. M. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays
using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. _J. Magn. Reson. Ser. B_ 108, 94–98 (1995). Article CAS Google Scholar * Takeuchi, K., Heffron, G., Sun, Z. Y.
J., Frueh, D. P. & Wagner, G. Nitrogen-Detected CAN and CON experiments as alternative experiments for main chain NMR resonance assignments. _J. Biomol. NMR_ 47, 271–282 (2010). Article
CAS PubMed PubMed Central Google Scholar * Pontoriero, L. et al. NMR reveals specific tracts within the intrinsically disordered regions of the SARS-CoV-2 nucleocapsid protein involved
in RNA encountering. _Biomolecules_ 12, 929 (2022). Article CAS PubMed PubMed Central Google Scholar * Schiavina, M., Pontoriero, L., Tagliaferro, G., Pierattelli, R. & Felli, I.
C. The role of disordered regions in orchestrating the properties of multidomain proteins: the SARS-CoV-2 nucleocapsid protein and its interaction with enoxaparin. _Biomolecules_ 12, 1302
(2022). Article CAS PubMed PubMed Central Google Scholar * Emsley, L. & Bodenhausen, G. Gaussian pulse cascades: new analytical functions for rectangular selective inversion and
in-phase excitation in NMR. _Chem. Phys. Lett._ 165, 469–476 (1990). Article CAS ADS Google Scholar * Emsley, L. & Bodenhausen, G. Optimization of shaped selective pulses for NMR
using a quaternion description of their overall propagators. _J. Magn. Reson._ 97, 135–148 (1992). CAS ADS Google Scholar * Slad, S., Bermel, W., Kümmerle, R., Mathieu, D. & Luy, B.
Band-selective universal 90° and 180° rotation pulses covering the aliphatic carbon chemical shift range for triple resonance experiments on 1.2 GHz spectrometers. _J. Biomol. NMR_ 76,
185–195 (2022). Article CAS PubMed PubMed Central Google Scholar * Khaneja, N., Reiss, T., Kehlet, C., Schulte-Herbrüggen, T. & Glaser, S. J. Optimal control of coupled spin
dynamics: design of NMR pulse sequences by gradient ascent algorithms. _J. Magn. Reson._ 172, 296–305 (2005). Article CAS PubMed ADS Google Scholar * Geen, H. & Freeman, R.
Band-selective radiofrequency pulses. _J. Magn. Reson._ 93, 93–141 (1991). ADS Google Scholar * Klika, K. D. The application of simple and easy to implement decoupling pulse scheme
combinations to effect decoupling of large J values with reduced artifacts. _Int. J. Spectrosc._ 2014, 1–9 (2014). Article Google Scholar * Garwood, M. & DelaBarre, L. The return of
the frequency sweep: designing adiabatic pulses for contemporary NMR. _J. Magn. Reson._ 153, 155–177 (2001). Article CAS PubMed ADS Google Scholar * Kelly, A. E., Ou, H. D., Withers, R.
& Dötsch, V. Low-conductivity buffers for high-sensitivity NMR measurements. _J. Am. Chem. Soc._ 124, 12013–12019 (2002). Article CAS PubMed Google Scholar * Voehler, M. W.,
Collier, G., Young, J. K., Stone, M. P. & Germann, M. W. Performance of cryogenic probes as a function of ionic strength and sample tube geometry. _J. Magn. Reson._ 183, 102–109 (2006).
Article CAS PubMed PubMed Central ADS Google Scholar * de Swiet, T. M. Optimal electric fields for different sample shapes in high resolution NMR spectroscopy. _J. Magn. Reson._ 174,
331–334 (2005). Article PubMed ADS Google Scholar * Takeda, M. et al. Construction and performance of an nmr tube with a sample cavity formed within magnetic susceptibility-matched
glass. _J. Magn. Reson._ 209, 167–173 (2011). Article CAS PubMed PubMed Central ADS Google Scholar * Bohlen, J.-M., Rey, M. & Bodenhausen, G. Refocusing with chirped pulses for
broadband excitation without phase dispersion. _J. Magn. Reson._ 84, 191–197 (1989). ADS Google Scholar * Kupce, E. & Freeman, R. Polychromatic selective pulses. _J. Magn. Reson. Ser.
A_ 102, 122–126 (1993). Article CAS ADS Google Scholar * Kupce, E., Boyd, J. & Campbell, I. D. Short selective pulses for biochemical applications. _J. Magn. Reson. Ser. B_ 106,
300–303 (1995). Article CAS Google Scholar * Smith, M. A., Hu, H. & Shaka, A. J. Improved broadband inversion performance for NMR in liquids. _J. Magn. Reson._ 151, 269–283 (2001).
Article CAS ADS Google Scholar * Claridge, T. D. W. High-Resolution NMR Techniques in Organic Chemistry 3rd ed. (Elsevier, 2016). * Shaka, A. J., Keeler, J. & Freeman, R. Evaluation
of a new broadband decoupling sequence: WALTZ-16. _J. Magn. Reson._ 53, 313–340 (1983). CAS ADS Google Scholar * Shaka, A. J., Barker, P. B. & Freeman, R. Computer-optimized
decoupling scheme for wideband applications and low-level operation. _J. Magn. Reson._ 64, 547–552 (1985). CAS ADS Google Scholar * Kadkhodaie, M., Rivas, O., Tan, M., Mohebbi, A. &
Shaka, A. Broadband homonuclear cross polarization using flip-flop spectroscopy. _J. Magn. Reson._ 91, 437–443 (1991). CAS ADS Google Scholar * Markley, J. L. et al. Recommendations for
the presentation of NMR structures of proteins and nucleic acids. _Pure Appl. Chem._ 70, 117–142 (1998). Article CAS Google Scholar * Hsu, S.-T. D., Bertoncini, C. W. & Dobson, C. M.
Use of protonless NMR spectroscopy to alleviate the loss of information resulting from exchange-broadening. _J. Am. Chem. Soc._ 131, 7222–7223 (2009). Article CAS PubMed Google Scholar *
Nováček, J. et al. 5D 13C-detected experiments for backbone assignment of unstructured proteins with a very low signal dispersion. _J. Biomol. NMR_ 50, 1–11 (2011). Article PubMed Google
Scholar * Pantoja-Uceda, D. & Santoro, J. Direct correlation of consecutive C′–N groups in proteins: a method for the assignment of intrinsically disordered proteins. _J. Biomol. NMR_
57, 57–63 (2013). Article CAS PubMed Google Scholar * Lopez, J., Schneider, R., Cantrelle, F., Huvent, I. & Lippens, G. Studying intrinsically disordered proteins under true in vivo
conditions by combined cross-polarization and carbonyl-detection NMR spectroscopy. _Angew. Chem. Int. Ed._ 128, 7544–7548 (2016). Article ADS Google Scholar * Cook, E. C., Usher, G. A.
& Showalter, S. A. The use of 13C direct-detect NMR to characterize flexible and disordered proteins. _Methods Enzymol._ 611, 81–100 (2018). Article CAS PubMed Google Scholar * Alik,
A. et al. Sensitivity‐enhanced 13C NMR spectroscopy for monitoring multisite phosphorylation at physiological temperature and pH. _Angew. Chem. Int. Ed._ 59, 10411–10415 (2020). Article
CAS Google Scholar * Ozenne, V. et al. Flexible-Meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental
observables. _Bioinformatics_ 28, 1463–1470 (2012). Article CAS PubMed Google Scholar * Brutscher, B. et al. in _Intrinsically Disordered Proteins Studied by NMR Spectroscopy_ (eds.
Felli, C. I. & Pierattelli, R.) 49–122 (Springer, 2015). * Pontoriero, L., Schiavina, M., Murrali, M. G., Pierattelli, R. & Felli, I. C. Monitoring the interaction of Α‐synuclein
with calcium ions through exclusively heteronuclear nuclear magnetic resonance experiments. _Angew. Chem. Int. Ed._ 59, 18537–18545 (2020). Article CAS Google Scholar * Schanda, P.,
Forge, V. & Brutscher, B. HET-SOFAST NMR for fast detection of structural compactness and heterogeneity along polypeptide chains. _Magn. Reson. Chem._ 44, 177–184 (2006). Article Google
Scholar * Schanda, P., Van Melckebeke, H. & Brutscher, B. Speeding up three-dimensional protein NMR experiments to a few minutes. _J. Am. Chem. Soc._ 128, 9042–9043 (2006). Article
CAS PubMed Google Scholar * Hošek, T., Gil-Caballero, S., Pierattelli, R., Brutscher, B. & Felli, I. C. Longitudinal relaxation properties Of1HN and 1Hα determined by direct-detected
13C NMR experiments to study intrinsically disordered proteins (IDPs). _J. Magn. Reson._ 254, 19–26 (2015). Article PubMed ADS Google Scholar * Bermel, W. et al. Speeding up sequence
specific assignment of IDPs. _J. Biomol. NMR_ 53, 293–301 (2012). Article CAS PubMed Google Scholar * Lescop, E., Schanda, P. & Brutscher, B. A set of BEST triple-resonance
experiments for time-optimized protein resonance assignment. _J. Magn. Reson._ 187, 163–169 (2007). Article CAS PubMed ADS Google Scholar * Chen, J.-H. & Mao, X.-A. Radiation
damping transfer in nuclear magnetic resonance experiments via chemical exchange. _J. Chem. Phys._ 107, 7120–7126 (1997). Article CAS ADS Google Scholar * Krishnan, V. V. & Murali,
N. Radiation damping in modern NMR experiments: progress and challenges. _Prog. Nucl. Magn. Reson. Spectrosc._ 68, 41–57 (2013). Article CAS PubMed Google Scholar Download references
ACKNOWLEDGEMENTS This paper is part of a project funded by the European Union-NextGenerationEU through the ItaliaDomani PNRR project ‘Potentiating the Italian Capacity for Structural Biology
Services in Instruct-ERIC’ (ITACA.SB, no. IR0000009). The support of the CERM/CIRMMP center of Instruct-ERIC and of the Italian Ministry for University and Research (MUR, FOE funding) is
gratefully acknowledged. MUR and Bruker Switzerland AG are acknowledged for financial support to M.A.R. (DM 352/2022) and MUR for financial support to L.B. (Dipartimenti di Eccellenza
2018-2022). Further support has been provided by the ItaliaDomani PNRR projects ‘Tuscany Health Ecosystem’ (THE, no. ECS00000017) and ‘A multiscale integrated approach to the study of the
nervous system in health and disease’ (MNESYS, no. PE0000006). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Chemistry ‘Ugo Schiff’ and Magnetic Resonance Center (CERM),
University of Florence, Florence, Italy Marco Schiavina, Lorenzo Bracaglia, Maria Anna Rodella, Isabella C. Felli & Roberta Pierattelli * Bruker BioSpin AG, Fällanden, Switzerland Rainer
Kümmerle * Department of Computational and Structural Biology, Max Perutz Labs, University of Vienna, Vienna, Austria Robert Konrat Authors * Marco Schiavina View author publications You
can also search for this author inPubMed Google Scholar * Lorenzo Bracaglia View author publications You can also search for this author inPubMed Google Scholar * Maria Anna Rodella View
author publications You can also search for this author inPubMed Google Scholar * Rainer Kümmerle View author publications You can also search for this author inPubMed Google Scholar *
Robert Konrat View author publications You can also search for this author inPubMed Google Scholar * Isabella C. Felli View author publications You can also search for this author inPubMed
Google Scholar * Roberta Pierattelli View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.S., I.C.F. and R.P. conceived and designed the
protocol. All authors contributed to the NMR experiments. M.S., L.B. and M.A.R. analyzed the data. All authors wrote, read and commented on the paper. CORRESPONDING AUTHORS Correspondence to
Marco Schiavina, Isabella C. Felli or Roberta Pierattelli. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature
Protocols_ thanks Davy Sinnaeve and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. RELATED LINKS KEY REFERENCES USING THIS PROTOCOL Pontoriero, L. et al. _Angew. Chem. Int. Ed_.
59, 18537–18545 (2020): https://doi.org/10.1002/anie.202008079 Schiavina, M. et al. _Biophys J_. 117, 46–55 (2019): https://doi.org/10.1016/j.bpj.2019.05.017 Murrali, M. G. et al.
_Chembiochem_. 19, 1625–1629 (2019) https://doi.org/10.1002/cbic.201800172 Banci, L. et al. GHz. Preprint at _arXiv_ (2019): https://doi.org/10.48550/arXiv.1910.07462 SUPPLEMENTARY
INFORMATION REPORTING SUMMARY RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement
with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and
applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Schiavina, M., Bracaglia, L., Rodella, M.A. _et al._ Optimal 13C NMR investigation of intrinsically disordered
proteins at 1.2 GHz. _Nat Protoc_ 19, 406–440 (2024). https://doi.org/10.1038/s41596-023-00921-9 Download citation * Received: 03 April 2023 * Accepted: 20 September 2023 * Published: 12
December 2023 * Issue Date: February 2024 * DOI: https://doi.org/10.1038/s41596-023-00921-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
