Establishment of a chinese critical care database from electronic healthcare records in a tertiary care medical center
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT The medical specialty of critical care, or intensive care, provides emergency medical care to patients suffering from life-threatening complications and injuries. The medical
specialty is featured by the generation of a huge amount of high-granularity data in routine practice. Currently, these data are well archived in the hospital information system for the
primary purpose of routine clinical practice. However, data scientists have noticed that in-depth mining of such big data may provide insights into the pathophysiology of underlying diseases
and healthcare practices. There have been several openly accessible critical care databases being established, which have generated hundreds of scientific outputs published in scientific
journals. However, such work is still in its infancy in China. China is a large country with a huge patient population, contributing to the generation of large healthcare databases in
hospitals. In this data descriptor article, we report the establishment of an openly accessible critical care database generated from the hospital information system. SIMILAR CONTENT BEING
VIEWED BY OTHERS HARNESSING BIG DATA IN CRITICAL CARE: EXPLORING A NEW EUROPEAN DATASET Article Open access 28 March 2024 UTILIZING BIG DATA FROM ELECTRONIC HEALTH RECORDS IN PEDIATRIC
CLINICAL CARE Article 24 November 2022 SALZBURG INTENSIVE CARE DATABASE (SICDB): A DETAILED EXPLORATION AND COMPARATIVE ANALYSIS WITH MIMIC-IV Article Open access 20 May 2024 BACKGROUND
& SUMMARY Critically ill patients managed in the intensive care unit (ICU) are usually monitored closely for organ dysfunctions, and are treated intensively by a variety of supportive
modalities1,2. Vital signs, laboratory tests, and medical treatments were obtained at a higher frequency than those treated in the general ward. Such daily intensive management will produce
a huge amount of information including medical orders, imaging studies, laboratory findings, and waveform signals. The data generation mechanisms may reflect key factors related to the
healthcare system, the pathophysiology of underlying disease, and patient’s preferences and cultures3. Thus, in-depth data mining of such large databases, such as risk factor analysis,
predictive analytics, and causal inference4,5,6, can provide more insights into clinical research questions. More knowledge or pearls of wisdom can be obtained from data mining, and the
translation of the knowledge into clinical practice may potentially improve clinical outcomes7,8. Most published scientific reports do not make their original raw data freely accessible in
the current critical care research community, partly attributable to confidentiality issues. The unwillingness to share data makes it difficult to reproduce the reported results.
Furthermore, the exploration of a such large database from a single research group could be biased and limited. Thus, strenuous efforts have been made to encourage the scientific community
to share their raw data, which is also supported by the open data campaign9,10. Several openly accessible critical care databases have been established, mainly reflecting the healthcare
systems of western countries11,12,13. China is a large country with a huge patient population. For example, the estimated incident sepsis cases are about 3 million in 2017, accounting for
nearly 10% of the global incident cases14. Chinese hospitals also have special hospital information systems that are distinct from those of western countries. However, hospital information
systems in Chinese hospitals are mainly used for clinical practice and are far less developed for research purposes. Data sharing is still in its infancy in the Chinese critical care
community, which significantly impairs the transparency of scientific work and international collaborations. To the best of our knowledge, there are two critical care databases being
established in China which focus on pediatric critically ill patients and those with infections15,16. Here, we reported the establishment of a large critical care database comprising
high-granularity data generated from the information system of a tertiary care university hospital. Details of the database are reported in the paper to encourage new research through
secondary analysis of the database. METHODS STUDY SETTING AND POPULATION The study was conducted in Zhejiang Provincial People’s Hospital, Zhejiang, China from January 2012 to May 2022. All
patients admitted to the ICU of the hospital were eligible. There were two ICUs in the hospital: one was the comprehensive central ICU and the other was the emergency ICU (EICU). There was
no exclusion criterion in enrolling subjects because we believed that patients who were excluded by a particular study might be eligible for another study. Thus, we included all records in
the information system related to ICU stays. The study was approved by the ethics committee of Zhejiang Provincial People’s Hospital (approval number: QT2022185). Informed consent was waived
as determined by the institutional review board, due to the retrospective design of the study. The study was conducted in accordance with the Declaration of Helsinki. DATABASE STRUCTURE AND
DEVELOPMENT The database is distributed as comma-separated value (CSV) files that can be imported to any relational database system. Each file contains a single table which will be further
explained in the subsequent sections. Each individual subject can be identified by a series number (patient_SN) with the combination of digits and letters such as
“3c74cf74c36241b7082ec35e458279dc”. Each unit hospital stay is denoted by a _Hospital_ID_ with examples such as “9432117” and “336688072433”. The unique ICU stay can be identified by the
_HospitalTransfer_ table, which contains intrahospital transfer events for the subjects. All tables use _Hospital_ID_ to identify an individual hospital stay, and the _HospitalTransfer_
table can be used to determine ICU stays linked to the same patient and/or hospitalization. We recommend the R package _tidyverse_ for the management of the relational database because of
its capability to streamline the workflow from data management to statistical analysis and to the training of machine learning models17. For large files, we recommend the _data.table_
package to process the tabular data. DEIDENTIFICATION All tables are deidentified according to the Health Insurance Portability and Accountability Act (HIPAA). All protected information is
removed including addresses, date of birth, date of hospital admission, date of discharge, date of medical order, personal numbers (e.g. name, phone, social security, and hospital number),
exact age on admission (age is discretized into bins). When creating the dataset, patients were randomly assigned a unique identifier (patient_SN and hospital_ID) and the original hospital
identifiers were not retained. As a result, the identifiers in the database cannot be linked back to the original, identifiable data. All doctor/nurse/pharmacist identifiers have also been
removed to protect the privacy of contributing providers. DATA RECORDS The database comprises 8180 unique hospital admissions for 7638 individual patients from January 2012 to May 2022 and
is available at the PhysioNet repository18. Table 1 shows the baseline demographics of hospital admissions. There are 2965 female and 5215 male patients in the dataset. The length of
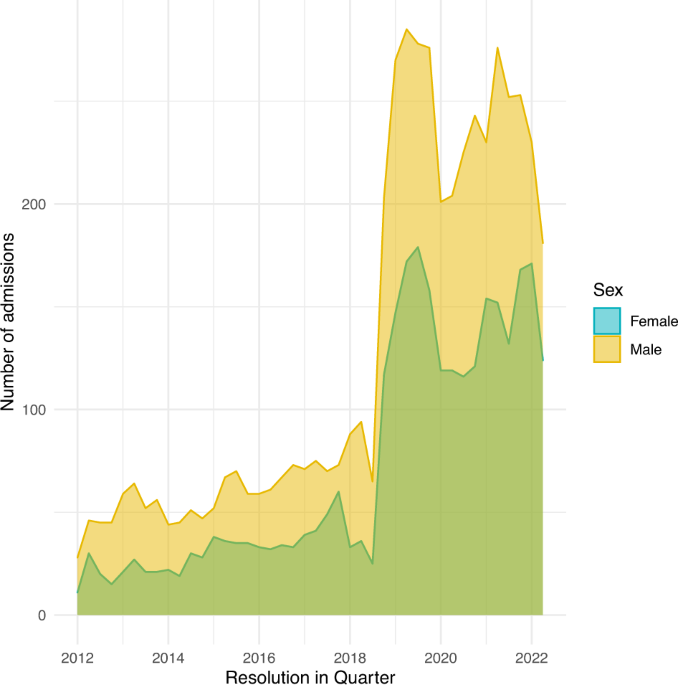
hospital days was 17 days (Q1 to Q3: 10 to 28). Male patients showed slightly longer hospital stay. The number of hospital admissions for ICU patients increased remarkably after the year
2018 because of the expansion of bed numbers this year for both comprehensive ICU and emergency ICU (Fig. 1). The distributions of hospital length of stay are shown in Fig. 2, restricting to
patients with a length of stay (LOS) <60 days. We then categorized specific diagnoses into 31 categories to explore the characteristics of the population in the dataset19. The
co-occurrences of the diseases are shown in Fig. 3. The results showed that pulmonary diseases are among the most common reasons for admission, followed by chronic heart failure (CHF). CHF
usually coexists with valvular disorders. It is also noted that pulmonary diseases usually coexist with cardiac arrhythmia (Fig. 3). Figure 4 shows the frequency of these diseases.
Hypertension is among the highest diseases in the study population, followed by chronic heart failure and arrhythmia. CLASSES OF DATA The data are organized into tables. There are a total of
17 tables comprising patient demographic data, medical order, laboratory findings, image studies, microbiology and hospital transfer events (Table 2). We will provide more details for each
individual table to promote the reuse of our database. PATIENT ADMISSION RECORD TABLE The patient admission record table describes the baseline patient demographics, past history, chief
complain, and length of stay in the hospital. The _patient_SN_ is a unique ID for individual patient and _Hospital_ID_ is unique ID for hospital admission. If a patient discharged/died
within 24 hours, the data were recorded in a separate table, so there are separate columns describing the chief complain and admission status for those short hospital stays. We provide both
English and Chinese descriptions for chief complain. The present history recorded in the _Med_history_ column contains more words, and the original Chinese descriptions are kept so that some
natural language processing algorithms can be applied. The StatusOnDischarge variable includes several categories such as Cured, Not cured, Unknown and Dead. These categories are recorded
as that in the original electronic system. The “Not cured” status refers to the situation when a patient was discharged against medical order and might be transferred to the primary care
service center or go home for palliative care. The “Unknown” label is also entered by the clinicians and should be considered as a separate type of status (Table 3). ELECTRONIC MEDICAL
RECORD (FIRST NOTE TABLE) The FirstNote.csv table contains data related to the progress note recorded on the admission day (Table 4), which includes free text data such as the reasons for
diagnosis, differential diagnosis and care plan. The diagnosis in this table is the initial diagnosis made on the day of admission and is subject modifications. PROGRESS NOTE TABLE The
progress note table (ProgressNote.csv) contains information on a variety of daily progress notes such as Daily course record, Blood transfusion record, and record for bedside procedures
(Table 5). DIAGNOSIS TABLE The diagnosis table contains information related to diagnosis for a hospital stay (Table 6). The _Diagnosis_Desc_ column provides free text description for the
diagnosis. ICD10_code is the code number for the standard ICD code. The information can be well processed with the _icd_ package in R (https://github.com/cran/icd). The functionality of the
package includes but not limited to finding comorbidities of patients based on ICD-10 codes, Charlson and Van Walraven score calculations, and comprehensive test suite to increase confidence
in accurate processing of ICD codes. HOSPITAL TRANSFER TABLE The _HospitalTransfer_ table contains information related to intrahospital transfer events (Table 7). The time and department of
each transfer event are given in respective columns. In the table, one row represents one transfer event, including the department a patient leaves (_TransferFrom_Dept_Eng_) and another
department a patient transfer into (_TransferTo_Dept_Eng_). One episode of hospitalization may contain multiple transfer events. To protect patients’ privacy, all date and time information
is recorded as days relative to hospital admission. Since the EICU is in the emergency department, the department names denoted by “Emergency medical department” or “Emergency Department”
refer to the EICU. SURGERY INFORMATION TABLE The surgical operation information is recorded in a separate table (SurgeryTab.csv). The table records the scheduled time for operation and
descriptions for the operation. The name of the operation can be extracted from the text descriptions (Oper_Scheduled). The medical order for a planned operation is usually prescribed 1 day
prior to the operation. If the planned date takes a minus value, it can be regarded that the operation is performed on the day of hospital admission (Table 8). THE LAB TABLE The lab table
contains data related to the laboratory findings (Table 9). There are 11,082,482 records of laboratory items in the dataset involving 214 types of laboratory items. there are 17 types of
samples being tested for laboratory findings, including whole blood, plasma, urine, serum, arterial blood, stool, venous blood, catheter orifice, ascites, bile, dialysate, CK blood sample
(kaolin-activated TEG channel), cerebrospinal fluid, bone marrow, deep venous catheter, sputum, gastric juice. The sample collection time is also recorded in days in reference to the
hospital admission time. The _Lab_category_ column may contain missing values for the following reasons: (1) the laboratory category is missing for some laboratory items that are derived
from other values, such as INR, Urea: creatinine, and Arterial alveolar oxygen partial pressure ratio; (2) Some laboratory items are exported from the bedside point-of-care machines, such as
troponin and blood gas items in an acute care setting; their laboratory category is not integrated into the laboratory system; and (3) some values not directly assayed by the machine such
as inspired oxygen saturation (FiO2), and prothrombin time control. Since the missing information in the laboratory category will not influence the research outcome; we did not populate
these missing cells. THE LAB DICTIONARY To facilitate the use of the Lab table, we generated a lab dictionary table (Table 10) which included the unique names of lab items and the lab
category. MICROBIOLOGY CULTURE TABLE The _MicrobiologyCulture_ table contains information related to microbiology culture results (Table 11). Conventional information regarding sample,
culture finding, culture time and description of microbiology culture are provided in the table. DRUG SENSITIVITY TABLE The _DrugSens_ table contains information related to the drug
sensitivity of cultured bacteria (Table 12). Conventional information including sample, microbiology, culture time, and drug name is available in the table. The negative and positive values
in the _DrugSens_result_ column refer to the results for Ultra broad spectrum β- Lactamase or D-test. EXAMINATION REPORT TABLE The _ExamReport_ table contains information related to a
variety of medical examinations, including computed topography (CT), X-ray and ultrasound (Table 13). The images are not available in current dataset, but instead we include the free text
descriptions and conclusions for these examinations. MEDICAL ORDER TABLE The _MedOrder_ table contains information related to the medical order prescribed by clinicians (Table 14). The table
provides both regular and stat medical orders (_MedOrder_Type_). The contents of the medical order can be found in the _MedOrder_DESC_ column. MEDICATION TABLE The medication table provides
data on the medication orders prescribed by clinicians (Table 15). This table is designed specifically for medication orders, containing columns for drug dose, frequency, unit of drug dose
and route of administration. MEDICATION DICTIONARY The Medication_Dictionary table provides information for the unique medication names. Some medications can be easily obtained from the
dictionary table. We provided a DrugName column where users can easily look up unified pharmaceutical names irrespective of the specifications, formula, and route of administration. For
example, if we want to extract sodium chloride injection, we can look for sodium chloride in the DrugName column. Alternatively, users may search the Med_DESC_Eng column with the key words
“Sodium chloride”. This can be easily achieved by the _stringr_ pipeline in R (Table 16). VITAL SIGN TABLE The _VitalSign_ table provides vital sign data for each hospital admission (Table
17). The _VitalSign_DESC_ column provides categories of vital signs including diastolic blood pressure, temperature, heart rate and respiratory rate. TECHNICAL VALIDATION Data were verified
for integrity during the data transfer process from the hospital information system to the database platform using MD5 checksums (Table 2). The MD5_hashes presented in Table 2 can also be
used by users to ensure the integrity of the downloaded datasets. All text information extracted from our medical information system are in Chinese. In establishing our data warehouse, we
translated some meta-data and short text to facilitate the use of data by researchers outside China. The translation was first performed by using the paid BaiDu academic translation service
(service number: MPE2022102608424528825) and then checked by two authors (Senjun Jin and Zhongheng Zhang) of the project. However, in order to maintain data fidelity, very little
post-processing has been performed for other long text fields such as present history, progress notes, and text reports of image studies. Some natural language contents were not translated
into English because any translations may change the results of natural language processing or text mining20,21. Users can employ some academic language translation services (including API)
for a large volume of language translation if needed. The medical data archived within the database were originally not intended for secondary analysis. Thus, some missing values and
inconsistencies may occur due to technical errors, system integration, and data preprocessing. In particular, the electronic critical care nursing chart system was launched in the year 2018,
and thus the current database contained no information before that time. These older nursing chart data before 2018 are recorded manually and archived in paper documents. We are planning to
convert these data into electronic information in a future project. USAGE NOTES DATA ACCESS Data are deposited in the PhysioNet repository (https://physionet.org/) and can be accessed after
completion of an online course (e.g. from the Collaborative Institutional Training Initiative)22. Data access also requires a data use agreement to be signed, which stipulates that the user
will not try to re-identify any subjects, will not share the data, and will release code associated with any publication using the data. Once approved, the plain CSV files can be directly
downloaded from the project on PhysioNet22. CODE AVAILABILITY The code for establishing the database was available on GitHub:
https://github.com/zh-zhang1984/ZhejiangProvinceICU/blob/main/ZhejiangProvinceICU.md REFERENCES * Elias, K. M., Moromizato, T., Gibbons, F. K. & Christopher, K. B. Derivation and
validation of the acute organ failure score to predict outcome in critically ill patients: a cohort study. _Crit Care Med_ 43, 856–864 (2015). Article Google Scholar * Yehya, N. &
Wong, H. R. Adaptation of a Biomarker-Based Sepsis Mortality Risk Stratification Tool for Pediatric Acute Respiratory Distress Syndrome. _Crit Care Med_ 46, e9–e16 (2018). Article Google
Scholar * Chu, C. D. _et al_. Trends in Chronic Kidney Disease Care in the US by Race and Ethnicity, 2012–2019. _JAMA Netw Open_ 4, e2127014 (2021). Article Google Scholar * Höfler, M.
Causal inference based on counterfactuals. _BMC Med Res Methodol_ 5, 28 (2005). Article Google Scholar * Zhang, Z., Chen, L., Xu, P. & Hong, Y. Predictive analytics with ensemble
modeling in laparoscopic surgery: A technical note. _Laparoscopic, Endoscopic and Robotic Surgery_ https://doi.org/10.1016/j.lers.2021.12.003 (2022). Article Google Scholar * Zhang, Z. _et
al_. Causal inference with marginal structural modeling for longitudinal data in laparoscopic surgery: A technical note. _Laparoscopic, Endoscopic and Robotic Surgery_
https://doi.org/10.1016/j.lers.2022.10.002 (2022). Article Google Scholar * Valik, J. K. _et al_. Validation of automated sepsis surveillance based on the Sepsis-3 clinical criteria
against physician record review in a general hospital population: observational study using electronic health records data. _BMJ Qual Saf_ 29, 735–745 (2020). Article Google Scholar *
Zhang, Z. _et al_. Analytics with artificial intelligence to advance the treatment of acute respiratory distress syndrome. _J Evid Based Med_ 13, 301–312 (2020). Article Google Scholar *
Forero, D. A., Curioso, W. H. & Patrinos, G. P. The importance of adherence to international standards for depositing open data in public repositories. _BMC Res Notes_ 14, 405 (2021).
Article Google Scholar * Shahin, M. H. _et al_. Open Data Revolution in Clinical Research: Opportunities and Challenges. _Clin Transl Sci_ 13, 665–674 (2020). Article Google Scholar *
Pollard, T. J. _et al_. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. _Sci Data_ 5, 180178 (2018). Article Google Scholar *
Johnson, A. E. W. _et al_. MIMIC-III, a freely accessible critical care database. _Sci Data_ 3, 160035 (2016). Article CAS Google Scholar * Thoral, P. J. _et al_. Sharing ICU Patient
Data Responsibly Under the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration: The Amsterdam University Medical Centers Database
(AmsterdamUMCdb) Example. _Crit Care Med_ 49, e563–e577 (2021). Article Google Scholar * Rudd, K. E. _et al_. Global, regional, and national sepsis incidence and mortality, 1990–2017:
analysis for the Global Burden of Disease Study. _Lancet_ 395, 200–211 (2020). Article Google Scholar * Zeng, X. _et al_. PIC, a paediatric-specific intensive care database. _Sci Data_ 7,
14 (2020). Article Google Scholar * Xu, P. _et al_. Critical Care Database Comprising Patients With Infection. _Front Public Health_ 10, 852410 (2022). Article Google Scholar * Wickham,
H. _et al_. Welcome to the Tidyverse. _Journal of Open Source Software_ 4, 1686 (2019). Article ADS Google Scholar * Jin, S., Chen, L., Chen, K. & Zhang, Z. Establishment of a Chinese
critical care database from electronic healthcare records in a tertiary care medical center (version 1.0). _PhysioNet_ https://doi.org/10.13026/3h21-rc35 (2022). * Quan, H. _et al_. Coding
algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. _Med Care_ 43, 1130–1139 (2005). Article Google Scholar * Li, S. _et al_. Deep Phenotyping of Chinese
Electronic Health Records by Recognizing Linguistic Patterns of Phenotypic Narratives With a Sequence Motif Discovery Tool: Algorithm Development and Validation. _J Med Internet Res_ 24,
e37213 (2022). Article Google Scholar * Gong, L., Zhang, Z. & Chen, S. Clinical Named Entity Recognition from Chinese Electronic Medical Records Based on Deep Learning Pretraining. _J
Healthc Eng_ 2020, 8829219 (2020). Article Google Scholar * Goldberger, A. L. _et al_. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex
physiologic signals. _Circulation_ 101, E215–220 (2000). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS S.J. received funding from Youth Talents Project of Health
Commission of Zhejiang Province (Project number: 2019RC103) and Health Science and Technology Plan of Zhejiang Province (2023KY051). Z.Z. received funding from Yilu “Gexin” - Fluid Therapy
Research Fund Project (YLGX-ZZ-2020005), Health Science and Technology Plan of Zhejiang Province (2021KY745). AUTHOR INFORMATION Author notes * These authors contributed equally: Senjun Jin,
Lin Chen, Chaozhou Hu, Sheng’an Hu. AUTHORS AND AFFILIATIONS * Emergency and Critical Care Center, Department of Emergency Medicine, Zhejiang Provincial People’s Hospital, Affiliated
People’s Hospital, Hangzhou Medical College, Hangzhou, Zhejiang, 310014, China Senjun Jin, Chaozhou Hu & Sheng’an Hu * Department of Critical Care Medicine, Affiliated Jinhua Hospital,
Zhejiang University School of Medicine, Jinhua, China Lin Chen & Kun Chen * Department of Emergency Medicine, Key Laboratory of Precision Medicine in Diagnosis and Monitoring Research of
Zhejiang Province, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310016, China Zhongheng Zhang Authors * Senjun Jin View author publications You can also
search for this author inPubMed Google Scholar * Lin Chen View author publications You can also search for this author inPubMed Google Scholar * Kun Chen View author publications You can
also search for this author inPubMed Google Scholar * Chaozhou Hu View author publications You can also search for this author inPubMed Google Scholar * Sheng’an Hu View author publications
You can also search for this author inPubMed Google Scholar * Zhongheng Zhang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS S.J. and Z.Z.
conceived the idea; L.C. and S.J. curated data; Y.H., H.C. and H.S. checked the accuracy of the data. CORRESPONDING AUTHOR Correspondence to Zhongheng Zhang. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and
institutional affiliations. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing,
adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons
license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a
credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted
use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT
THIS ARTICLE CITE THIS ARTICLE Jin, S., Chen, L., Chen, K. _et al._ Establishment of a Chinese critical care database from electronic healthcare records in a tertiary care medical center.
_Sci Data_ 10, 49 (2023). https://doi.org/10.1038/s41597-023-01952-3 Download citation * Received: 09 August 2022 * Accepted: 10 January 2023 * Published: 23 January 2023 * DOI:
https://doi.org/10.1038/s41597-023-01952-3 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
