Transcriptomic profiles of tumor-associated neutrophils reveal prominent roles in enhancing angiogenesis in liver tumorigenesis in zebrafish
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT We have previously demonstrated the pro-tumoral role of neutrophils using a _kras_-induced zebrafish hepatocarcinogenesis model. To further illustrate the molecular basis of the
pro-tumoral role, Tumor-associated neutrophils (TANs) were isolated by fluorescence-activated cell sorting (FACS) and transcriptomic analyses were carried out by RNA-Seq. Differentially
expressed gene profiles of TANs from larvae, male and female livers indicate great variations during liver tumorigenesis, but the common responsive canonical pathways included an immune
pathway (Acute Phase Response Signaling), a liver metabolism-related pathway (LXR/RXR Activation) and Thrombin Signaling. Consistent with the pro-tumoral role of TANs, gene module analysis
identified a consistent down-regulation of Cytotoxicity module, which may allow continued proliferation of malignant cells. Gene Set Enrichment Analysis indicated up-regulation of several
genes promoting angiogenesis. Consistent with this, we found decreased density of blood vessels accompanied with decreased oncogenic liver sizes in neutrophil-depleted larvae. Collectively,
our study has indicated some molecular mechanisms of the pro-tumoral roles of TANs in hepatocarcinogenesis, including weakened immune clearance against tumor cells and enhanced function in
angiogenesis. SIMILAR CONTENT BEING VIEWED BY OTHERS LIVER TUMOUR IMMUNE MICROENVIRONMENT SUBTYPES AND NEUTROPHIL HETEROGENEITY Article 09 November 2022 COMPARATIVE TRANSCRIPTOMICS COUPLED
TO DEVELOPMENTAL GRADING VIA TRANSGENIC ZEBRAFISH REPORTER STRAINS IDENTIFIES CONSERVED FEATURES IN NEUTROPHIL MATURATION Article Open access 27 February 2024 CANCER CELL GENETICS SHAPING OF
THE TUMOR MICROENVIRONMENT REVEALS MYELOID CELL-CENTRIC EXPLOITABLE VULNERABILITIES IN HEPATOCELLULAR CARCINOMA Article Open access 22 March 2024 INTRODUCTION Hepatocellular carcinoma (HCC)
is the most common type of primary liver cancer with high malignancy and mortality1. HCC is frequently caused by chronic inflammation in the liver, where the immune cells create an
unresolved, chronic inflammation by initiating and maintaining infiltrating immune cells and producing cytokines in the liver2,3. Among these immune cells, neutrophils, the most abundant
immune cells in human, have been proved to play a role in a variety of tumors4,5. In recent studies, various roles of tumor-associated neutrophils (TANs) have been identified, including the
existence of N1 (anti-tumoral) and N2 (pro-tumoral) tumor-associated neutrophils in tumor development and progression6. We have previously developed several inducible HCC models by
transgenic expression of selected driver oncogenes7,8,9,10,11. In these transgenic models, the driver oncogene can be temporally activated to initiate liver carcinogenesis and histologically
proven HCC are usually produced in a few weeks. Thus, these transgenic models provide a powerful tool for investigation of liver tumor initiation. In previous studies, we have found a
prominent role of immune response during liver cancer progression12. In particular, we found a rapid migration of neutrophils towards oncogenic liver in a _kras_-induced zebrafish HCC
model13. By manipulation of neutrophil numbers and activities through pharmaceutical treatments and genetic knockdown, we have demonstrated that tumor-associated neutrophils (TANs) promoted
hepatocarcinogenesis during the initiation of _kras_-induced oncogenesis. We found that increased TAN activity accelerated the proliferation of oncogenic hepatocytes and deterred their
apoptosis, thus contributing to the pathological malignancy. In that study, although the pro-tumor roles of TANs have been demonstrated, these roles remain to be elucidated at the molecular
level. The purpose of this study was to use a transcriptomic approach to provide molecular insights into TANs and their roles in hepatocarcinogenesis. Thus, neutrophils were isolated from
larvae, adult males and adult females following oncogenic _kras_ activation and used for RNA-seq analyses. By comparing transcriptomic profiles of TANs and matched naïve neutrophils (NNs),
we observed prominent roles of TANs in loss of cytotoxicity and pro-angiogenesis, both of which apparently favor tumor initiation and progression. RESULTS AND DISCUSSION VALIDATION OF
NEUTROPHIL IDENTITY BY TRANSCRIPTOMIC PROFILING TANs were isolated from _kras_+_/lyz_+ fish (see Methods) and NNs isolated from _lyz_+ fish by FACS. The fluorescent microscope images and
flow cytometry dot-plots of the isolated cells showed an obvious enrichment of DsRed+ neutrophils in comparison to the cell suspension before sorting (Supplementary Fig. S1). The purity of
the sorted neutrophils (DsRed+) is above 90%. The neutrophil samples from larva (L), male (M) and female (F) adult fish were collected with biological duplicates or triplicates, as
summarized in Table S1. RNAs were isolated from these neutrophil samples and sequenced to a depth of 23.4–98.9 × 106 reads for each library. In total, 15 RNA samples were sequenced,
including three TAN_L and three matched NN_L from larvae, three TAN_M and two NN_M from male adults, and two TAN_F and two NN_F from female adults. All the sequence reads were then mapped to
the zebrafish genome reference, danRer7, after removing low-quality reads. By annotating the mapped reads, a total of 8.8–13.5 × 103 transcript entries were identified with at least one
mapped read, constituting 60–90% of total known zebrafish transcript entries in the danRer7 zebrafish genome database (14,868 RefSeq transcript entries in total). To minimize the effect from
potentially leaky expression, a cut-off of 30 reads was used to retain the robustly expressed transcripts. As a result, 6.2–10.7 × 103 transcript entries were identified for each pool of
neutrophils and used for subsequent analyses (Supplementary Table S1), representing 42–72% of the danRer7 database. The distribution of transcript entries and total counts over different
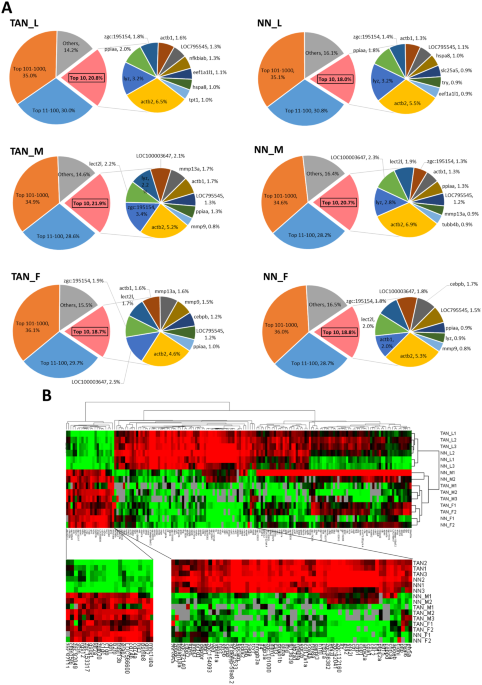
transcript abundance categories showed similar profiles for all neutrophil samples (Fig. 1A). There were only a few transcripts which had high abundance while the majority of transcripts
were at the very low abundance, which was the typical pattern of transcriptomic profiles of essentially all tissues and cell types14,15. The top 10 transcripts accounted for about 20%
transcriptome body and the top 100 transcripts constituted around 50% of the transcriptome body. In contrast, the lowest expressed ~11,000 transcript entries (ranked after 1000 and
categorized in others) contributed only about 15% of the transcriptome body. In the sub-pie of the top 10 abundant transcripts, the most abundant and common genes in all neutrophil groups
were _actb1, actb2, lyz_ and _lect2l_. Both _actb1_ and _actb2_ encodes β-actin, which are commonly known as housekeeping genes and play critical roles in cell mobility. Their crucial roles
in neutrophil functions have long been recognized as mutation of these genes in neutrophils could result in abnormalities in chemotaxis, superoxide production and membrane potential
response16. Both _lyz_ and _lect2l_ are well-known as neutrophil-specific genes. _lyz_ encodes a specific enzyme to hydrolyze specific linkages in bacterial cell wall17 while _lect21_ is a
leukocyte-derived chemotactic factor gene18. Both of them play significant roles against pathogen infection in neutrophils. Thus, these top abundantly expressed genes are consistent with the
function of neutrophils and validate the neutrophil identities of the cell population we isolated. To examine the similarities between the RNA-seq samples, we performed hierarchical
clustering across all the 15 samples based on gene expression abundance. These samples were clearly clustered into two branches, larva and adult (Fig. 1B). Thus, there was an overwhelming
influence of developmental stages on neutrophil transcriptomes. Two distinct gene clusters were enriched in larvae and adults respectively. Among the larva enriched genes, a number of
neutrophil developmental regulators were identified, such as _etv5a, hyal6_, _ctrp, ctrl_ and _ctbp2_. Among them, c_tbp2_ encodes a C-terminal binding protein, which co-activates neutrophil
differentiation with a zinc finger transcription factor19, indicating an active development of neutrophils in the larval stage (8 dpf), in which the adaptive immune system has not been
fully developed20. In the adult enriched gene cluster, _mhc1uha, cd74a_ and _cd74b_, which encode major histocompatibility complex (MHC) molecules, may indicate the maturity of the adaptive
immune system in adult fish. In addition, _psmb8_ and _b2m_ are important components of MHC I molecules21,22 while _grn1, grn2, rgs12, and tnfsfl3b_ are associated with neutrophil responses
to chemokines and cytokines23,24. DISTINCT TRANSCRIPTOMES BETWEEN TUMOR ASSOCIATED AND NAÏVE NEUTROPHILS In order to extract specific transcriptomic features for TANs, DEGs were identified
by comparison of matched TANs and NNs. By using selection criteria of fold change >1.25 and p-value <0.05, DEGs were selected from the three TAN/NN groups. The numbers of up- and
down-regulated genes in the larva, male and female TANs are shown in Venn diagrams in Fig. 2 and the list of these genes are shown in Supplementary Tables S2–S4, including 619 up- and 564
down-regulated genes in larva TANs (Supplementary Table S2), 403 up- and 368 down-regulated genes in male TANs (Supplementary Table S3), and 581 up- and 387-down-regulated genes in female
TANs (Supplementary Table S4). However, deregulated genes from the three TAN groups had relatively small overlaps with only seven commonly up-regulated genes and eight commonly
down-regulated genes (Fig. 2), indicating that both developmental stages and genders affected the response of neutrophils greatly during hepatocarcinogenesis. This is consistent with a
previous study on gender difference in neutrophils in responses to cytokine stimulations and malignant growth25. Moreover, the aged neutrophils have also been demonstrated to be different
from the young ones in human26. To verify the dynamic range of gene expression, reverse-transcription quantitative PCR (RT-qPCR) was performed for the fifteen common DEGs. As shown in
Supplementary Fig. S2, there was a good correlation between the RNA-seq and RT-qPCR results for up- and down-regulation of these common DEGs (Supplementary Fig. S2). The seven commonly
up-regulated genes are involved in various biological activities, including regulation of G-protein signaling (_rgs3_), lipid transport (_pltp_), transcriptional regulation (_raraa_ and
_znf710a_) and vascularization (_lgals2b, naa35_ and _esm1_). Among these genes, _esm1_ has been reported to be associated with cancer and it is involved in cell survival, cell cycle
progression, migration, angiogenesis, invasion and epithelial-mesenchymal transition during tumor invasion in colorectal cancer27. The eight commonly down-regulated genes also have multiple
functional implications, including apoptosis (_dap1b_), phosphatase activity (_ctdsp2_) and chromatin-associated protein (_cbx7a_), ion binding and metal transition (_pter_ and _phf20b_),
translation (_dalrd3, eif4ebp3_), and antigen presenting and processing (_psme2_). Notably, _psme2_ encodes a proteinase in immunoproteasome, which plays a role in the processing of class I
MHC peptides28. The consistent down-regulation of this gene in all the three TAN groups might indicate their suppressed antigen presenting function during hepatocarcinogenesis. COMMONLY
UPREGULATED CANONICAL PATHWAYS IN TANS AS REVEALED BY IPA To further understand biological properties of TANs, we identified canonical pathways enriched in DEGs in TANs via IPA. As shown in
Fig. 3, the overlapping pathways among the three TAN groups were few. The common pathways enriched in all TAN groups were Acute Phase Response Signaling, LXR/RXR Activation, and Thrombin
Signaling. The first two pathways were highly robust and appeared to be highly significant (p-value < 0.001) at the top of the lists from all three TAN groups. Acute Phase Response
Signaling is associated with cytokine production for innate immune cells in response to stimuli, such as malignant growth29. LXR/RXR Activation is involved in lipid metabolism and it has
been reported to be activated in HCC30,31. It is interesting to note that LXR activation has been demonstrated to impair neutrophil motility in an infection model, and the inhibition of
chemokine-induced RhoA activation has been identified as a putative underlying mechanism32,33. The functional implication of this pathway is consistent with our earlier observation that TANs
becomes more stagnant after infiltrating into the tumor microenvironment13. In addition, Thrombin Signaling plays an enhancing role in cell adhesion and has long been found to promote tumor
in metastasis initiation34,35. Thus, these data revealed crucial responses of neutrophil towards the oncogenic transformation of hepatocytes and tend to indicate their pro-tumor roles.
IMPAIRMENT OF CYTOTOXICITY FUNCTION IN TANS AS REVEALED BY IMMUNE MODULE ANALYSIS To further characterize immune response in the three TAN groups, gene module analysis was conducted. All
gene modules significantly up- and down-regulated are shown in Fig. 4A. These gene modules revealed distinct enrichments in the three TAN group; however, Module 2.1 (Cytotoxicity) was
consistently down-regulated among the three TAN groups. In this module, TAN_L, TAN_M and TAN_F contained 18, 15 and 14 leading edge genes and 14 of them were common in all the three TANs
(Fig. 4B). The top down-regulated genes included tumor suppressors such as _GPR56, RARRES3_, and _GLCCI1. GPR56_ encodes a non-classical adhesion receptor and has been identified as a new
type of adhesion receptor that binds to extracellular matrix proteins36. Mediated by this transmembrane receptor, the cell adhesion has formed a critical inhibitory process during cancer
progression; for example, _GPR56_ has been reported to suppress tumor growth and metastasis in a melanoma model37. _RARRES3_ has been identified as a class II tumor suppressor in B cell
chronic lymphocytic leukemia and colorectal cancer38,39. It is also down-regulated in human HCC tumors and its overexpression in hepatoma cells promotes apoptosis40. _GLCCI1_ is an early
marker of glucocorticoid-induced apoptosis, involving in breast cancer and colorectal cancer41,42,43,44. Consistent with the roles of _RARRES3_ and _GLCCI1_ in apoptosis, we also observed
previously that TANs inhibited apoptosis of oncogenic hepatocytes in zebrafish and thus promoted hepatocarcinogenesis13. Thus, the transcriptomic data further support the pro-tumor role of
TANs by suppression of tumor-killing capability through inhibition of apoptosis. RESEMBLANCE OF ZEBRAFISH TAN SIGNATURE TO MOUSE TAN TRANSCRIPTOME To further validate the isolated zebrafish
TANs and their potential functional conservation across species, zebrafish TAN transcriptomic data were compared by GSEA with the transcriptomic data from mouse TANs, which were isolated
from _Kras_-driven melanoma and were so far the best matched TAN data for our _kras_-driven liver tumor in zebrafish45. The dataset also contained microarray data of NNs isolated from mouse
bone marrow and the granulocyte-like myeloid derived suppressor cells (G-MDSC) isolated from melanoma respectively as non-tumor control and parallel tumor associated granulocytes. The
up-regulated gene list of each zebrafish TAN group was used to represent its transcriptomic signature. As shown in Fig. 5A–F, based on normalized enrichment scores (NES) and false discovery
rate (FDR), the zebrafish TAN groups show significantly high resemblance to the mouse TANs when they were compared against both NNs and G-MDSC. Thus, there is a significant similarity
between zebrafish TAN transcriptomes and mouse TAN transcriptome, which are highly distinguishable from either NN or G-MDSC. To gain biological insights of the similarity between zebrafish
and mouse TANs, the top leading edge genes between them are presented in Fig. 5G and Supplementary Tables S5–S7, and their strong association with tumors were noted. The list contains
pro-angiogenic genes (_Lgals1, Lgals3bpb, Il1b, Vegfa_), pro-tumor cytokines (_Il1b, Il10_, and _Il10ra_), and tumor-related transcriptional factors (_Egr1_ and _Egr2_), indicating that
zebrafish and mouse TANs may have similar pro-tumor characteristics in these aspects. These example genes could be potential biomarkers for diagnosis and therapy. For example, _IL10_ and
_IL10RA_ have been utilized as potential therapeutic targets in a mouse HCC model, as the use of anti-IL10 and anti-IL10RA oligodeoxynucleotide in the treatment has enhanced the anti-tumor
activity of macrophages46. PRO-ANGIOGENIC ROLE OF TANS There was initial evidence suggesting that TANs can affect tumor angiogenesis47. However, the mechanism remains unclear. As shown in
Fig. 6A, we found that many pro-angiogenic genes were differentially expressed in all three TAN groups. These pro-angiogenic genes included _vegfa_ (vascular endothelial growth factor),
_il1b_ (pro-angiogenic interleukin), _itgb1_ (integrin), _mmp14_ and _mmp9_ (matrix metallopeptidases) and gene encoding other cell adhesion molecules (_ilgals1_ and _igals3bp_). The
overwhelming up-regulation of these pro-angiogenesis genes in TANs has indicated a prominent role of neutrophils in promoting tumor angiogenesis during tumor initiation. Notably, _lgals3bpb_
was found to be up-regulated in TANs from both mouse melanoma and early zebrafish liver tumorigenesis based on the leading edge genes of cross-species transcriptomic comparison between
zebrafish and mice (Fig. 5G). Interestingly, _LGALS3BP_ is also up-regulated in human HCC and cirrhosis tissue48. Lgals3bp extensively interacts with extracellular matrix components
including fibronectin and β1-integrin, both of which are up-regulated in oncogenic hepatocytes based on our unpublished hepatocyte transcriptomic data. This interaction involved in fibrosis
and angiogenesis in the development of malignant liver tumors49. To evaluate the effect of neutrophils on tumor angiogenesis, the number of neutrophils was manipulated by suppression of
neutrophil differentiation with MO_gcsfr50 in _kras_+_/fli_+ double transgenic embryos. The effects of these morpholino oligonucleotides have been previously validated in earlier reports50
as well as in our laboratory13,51. In this study, as shown in Fig. 6B,C, we found that the number of neutrophils in the liver region in MO_gcsfr injected larva was significantly lower than
that in the larvae injected with the control MO_SC morpholino. Next, we injected mopholino oligonucleotides into the _kras_+_/fli_+ and _kras−/fli_+ embryos at one-cell stage and analyzed
under a confocal microscope at 6 dpf with Dox induction from 3 dpf. As shown in Fig. 6D, in the MO_SC injected groups, blood vessels were obviously increased in _kras_+_/fli_+ larvae
(middle) compared to _kras−/fli_+ larvae after Dox induction (left). The increased blood vessels as well as increased liver size were confirmed by quantification of _fli_+ blood vessel area
(Fig. 6E) and the 2D liver size (Fig. 6F), which were consistent with our observation of increased blood vessels in the _Myc_ transgenic zebrafish model following the induction of
_Myc_-mediated liver tumorigenesis52. Thus, MO_gcsfr injection suppressed neutrophil differentiation and resulted in significant decreases in both the density of blood vessels and liver
size. These observations made at two different dpf (RNA-seq at 8 dpf and morpholino knockdown at 6 dpf) are consistent for a pro-angiogenic role of neutrophils in the initial stage of
hepatocarcinogenesis and should add more confidence on the conclusion, which is also consistent with a prominent pro-angiogenic role of TANs in hepatocarcinogenesis as previously suggested
for human HCC53. METHODS ZEBRAFISH HUSBANDRY AND INDUCTION OF LIVER TUMORS All zebrafish experiments were carried out in accordance with the recommendations in the Guide for the Care and Use
of Laboratory Animals of the National Institutes of Health and the protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the National University of Singapore
(Protocol Number: 096/12). Five transgenic lines were used in this study: _Tg_(_fabp10::rtTA2s-M2; TRE2::EGFP-kras__G12V__)_, shorted as _kras_+, which was generated using a Tet-On system to
have liver-specific expression of oncogenic krasG12V 7; _Tg_(_lyz::DsRed)_, shorted as _lyz_+. in which neutrophils are labeled with DsRed under the control of neutrophil-specific _lyz_
(lysozyme C) promotor54; _Tg_(_fli1a::GFP)_55 and _Tg_(_fli1a::RFP_)56 with GFP- and RFP-labeled blood vessels respectively, both of which are shorted as _fli_+ in this report; LiPan
transgenic zebrafish, _Tg_(_fabp10::DsRed; ela3l::EGFP_) with DsRed expression in the liver and EGFP expression in exocrine pancreas57. To induce the expression of KrasG12V-EGFP in _kras_+
fish, 20 µg/ml doxycycline (Dox; Sigma, D9891) was used for larvae from 3 day post fertilization (dpf) to 8 dpf and for adult fish from 6 month post fertilization for 5 days. ISOLATION OF
NEUTROPHILS Neutrophils were isolated from both 8-dpf larvae (n > 50 each sample) after removal of head and tail parts to enrich the liver portion) and adult livers (4–7 fish pooled for
each sample) through fluorescence-activated cell sorting (FACS) using a cell sorter (BD Aria) following a previously described protocol58. TANs were isolated based on DsRed expression cells
from Dox-treated kras+/lyz+ double transgenic fish and NNs were isolated based on DsRed expression from _lyz_+ larvae. The purity of FACS isolated neutrophils was above 90%, and the number
of cells collected from each sample was above 10,000. MORPHOLINO KNOCKDOWN AND CONFOCAL IMAGING Morpholino knockdown was performed on _kras_+_/fli_+ or _kras−/fli_+ as previously
described13. Two morpholino oligonucleotides were designed and synthesized by GeneTools (Philomath, OR). A previously validated morpholino oligonucleotides for reduction (MO_gcsfr,
5′-GAAGCACAAGCGAGACGGATGCCAT-3′)50 of neutrophil population, as well as a standard control morpholino (MO_SC, 5′-CCTCTTACCTCAGTTACAATTTATA-3′) that targeted a human beta-globin intron, were
used in this study. These morpholinos were injected into zebrafish embryos at one-cell stage and the larvae developed from injected embryos were imaged at 6 dpf using a confocal microscope
(Carl Zeiss LSM510) for examination of their angiogenesis in livers. Measurement of neutrophil number, liver sizes and blood vessels was performed using ImageJ as previously described9,59
and >20 larva for each group were analyzed in this study. The difference between comparing groups was evaluated by student’s t-test using Graphpad Prism 6 (statistical significance: *p
< 0.05, **p < 0.01, ***p < 0.001). RNA EXTRACTION AND LIBRARY PREPARATION Following FACS sorting, the viability and purity of isolated neutrophils was tested immediately once the
cell sorting was finished. Only the sorted samples with over 98% DAPI negative (living cells) and over 90% DsRed positive (neutrophil marker) were processed for the RNA extraction using the
RNeasy Micro Kit (Qiagen, 74004). The quality and quantity of isolated RNA were examined with Bioanalyzer by using Agilent RNA 6000 Pico Kit (Agilent Technologies, 5067-1513) and all RNA
samples in this study had RNA integrity values above 7. These RNAs were processed to NGS (next generation sequencing)-qualified cDNA by using SMARTer Ultra Low Input RNA Kit for Sequencing
(Clontech Laboratories, Inc., 634848). The cDNAs were examined with Bioanalyzer by using Agilent High Sensitivity DNA Kit (Agilent Technologies, 5067-4626). DNA shearing was performed on
Covaris AFA system and the resulting DNA was in the 200–500 bp range. The sheared cDNA was prepared for constructing multiplex sequencing libraries using NEB DNA Library Prep Kits (New
England Biolabs Inc., E7370L and E7335S). Each multiplex occupied one lane and was sequenced on Illumina HiSeq NGS platform with the sequencing read length of 100 bp. RNA-SEQ DATA PROCESSING
AND ANNOTATION Sequence reads were assembled by aligning to the zebrafish reference genome sequence (Danio rerio, UCSC version danRer7, 2010) using TopHat2.0
(http://ccb.jhu.edu/software/tophat/index.shtml). All the zebrafish genes were annotated to human and mouse orthologous/homologous genes by retrieving from the Genome Institute of Singapore
Zebrafish Annotation Database (http://giscompute.gis.a-star.edu.sg/~govind/unigene_db/) as previously described60. The quantification of reads mapped to each gene was performed using a
python package, HT-Seq61 with ambiguous and non-unique alignments depleted. RNA-SEQ DATA ANALYSES Based on sequence count of each gene, which has been normalized against the total sequence
counts, hierarchical clustering was performed using Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) across all neutrophil samples to examine their similarities.
Differentially expressed genes (DEGs) between TANs and NNs were identified by using DESeq2, an R package, which normalizes the count data and models with negative binomial distribution62,
with cut-off p-value at 0.05 and fold change at 1.25. The biological insights of the DEGs were mined by using IPA (Ingenuity Pathway Analysis, Qiagen). The fold change and p-value of DEGs
were input for the establishment of causal networks and the overlap p-value for each pathway was calculated using one-sided Fisher’s exact test. The overlap p-value < 0.05 was considered
to be significant. Gene module analysis was conducted based on coordinated gene expression pattern from multiple disease conditions according to Chaussabel _et al_.63. The comparison between
zebrafish TAN signatures (defined by significantly up-regulated genes against NNs) and mouse TAN microarray dataset (GSE43254) obtained from GEO (Gene Expression Omnibus) was performed by
Gene Set Enrichment Analysis (GSEA)64. The enrichment score of the signatures was estimated by using an empirical phenotype-based permutation test and statistical significance of enrichment
score was estimated by FDR < 0.25. REFERENCES * Davis, G. L. _et al_. Hepatocellular carcinoma: management of an increasingly common problem. _Proc (Bayl Univ Med Cent)_ 21, 266–80
(2008). Article Google Scholar * Matsuzaki, K. _et al_. Chronic inflammation associated with hepatitis C virus infection perturbs hepatic transforming growth factor beta signaling,
promoting cirrhosis and hepatocellular carcinoma. _Hepatology_ 46, 48–57 (2007). Article CAS Google Scholar * Weber, A., Boege, Y., Reisinger, F. & Heikenwalder, M. Chronic liver
inflammation and hepatocellular carcinoma: persistence matters. _Swiss Med Wkly_ 141, w13197 (2011). PubMed Google Scholar * Zivkovic, M. _et al_. Oxidative burst of neutrophils against
melanoma B16-F10. _Cancer Lett_ 246, 100–8 (2007). Article CAS Google Scholar * Bellocq, A. _et al_. Reactive Oxygen and Nitrogen Intermediates Increase Transforming Growth Factor– β 1
Release from Human Epithelial Alveolar Cells through Two Different Mechanisms. _American Journal of Respiratory Cell and Molecular Biology_ 21, 128–136 (1999). Article CAS Google Scholar
* Piccard, H., Muschel, R. J. & Opdenakker, G. On the dual roles and polarized phenotypes of neutrophils in tumor development and progression. _Critical reviews in oncology/hematology_
82, 296–309 (2012). Article CAS Google Scholar * Chew, T. W. _et al_. Crosstalk of Ras and Rho: activation of RhoA abates Kras-induced liver tumorigenesis in transgenic zebrafish models.
_Oncogene_ (2013). * Li, Z. _et al_. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. _J Hepatol_ 56, 419–25 (2012). Article CAS
Google Scholar * Li, Z. _et al_. A transgenic zebrafish liver tumor model with inducible Myc expression reveals conserved Myc signatures with mammalian liver tumors. _Dis Model Mech_ 6,
414–23 (2013). Article CAS Google Scholar * Nguyen, A. T. _et al_. An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. _Dis Model Mech_
5, 63–72 (2012). Article CAS Google Scholar * Sun, L., Nguyen, A. T., Spitsbergen, J. M. & Gong, Z. Myc-induced liver tumors in transgenic zebrafish can regress in tp53 null mutation.
_PLoS One_ 10, e0117249 (2015). Article Google Scholar * Li, Z. _et al_. Transcriptomic analysis of a transgenic zebrafish hepatocellular carcinoma model reveals a prominent role of
immune responses in tumour progression and regression. _Int J Cancer_ 135, 1564–73 (2014). Article CAS Google Scholar * Yan, C., Huo, X., Wang, S., Feng, Y. & Gong, Z. Stimulation of
hepatocarcinogenesis by neutrophils upon induction of oncogenic kras expression in transgenic zebrafish. _J Hepatol_ 63, 420–8 (2015). Article CAS Google Scholar * Hegedus, Z. _et al_.
Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. _Mol Immunol_ 46, 2918–30 (2009). Article CAS Google Scholar * Zheng, W. _et al_. Comparative
transcriptome analyses indicate molecular homology of zebrafish swimbladder and mammalian lung. _PLoS One_ 6, e24019 (2011). Article ADS CAS Google Scholar * Nunoi, H. _et al_. A
heterozygous mutation of beta-actin associated with neutrophil dysfunction and recurrent infection. _Proc Natl Acad Sci USA_ 96, 8693–8 (1999). Article ADS CAS Google Scholar * Yang,
C.-T. _et al_. Neutrophils Exert Protection in the Early Tuberculous Granuloma by Oxidative Killing of Mycobacteria Phagocytosed from Infected Macrophages. _Cell host & microbe_ 12,
301–312 (2012). Article CAS Google Scholar * Yamagoe, S. _et al_. Expression of a neutrophil chemotactic protein LECT2 in human hepatocytes revealed by immunochemical studies using
polyclonal and monoclonal antibodies to a recombinant LECT2. _Biochem Biophys Res Commun_ 237, 116–20 (1997). Article CAS Google Scholar * Dumortier, A., Kirstetter, P., Kastner, P. &
Chan, S. Ikaros regulates neutrophil differentiation. _Blood_ 101, 2219–2226 (2003). Article CAS Google Scholar * Lam, S. H., Chua, H. L., Gong, Z., Lam, T. J. & Sin, Y. M.
Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, _in situ_ hybridization and immunological study. _Dev Comp Immunol_ 28, 9–28 (2004).
Article CAS Google Scholar * Liu, Y. _et al_. Mutations in proteasome subunit beta type 8 cause chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature with
evidence of genetic and phenotypic heterogeneity. _Arthritis Rheum_ 64, 895–907 (2012). Article CAS Google Scholar * Zhang, X., Ding, L. & Sandford, A. J. Selection of reference genes
for gene expression studies in human neutrophils by real-time PCR. _BMC Molecular Biology_ 6, 4–4 (2005). Article Google Scholar * Avula, L. R. _et al_. Whole-genome microarray analysis
and functional characterization reveal distinct gene expression profiles and patterns in two mouse models of ileal inflammation. _BMC Genomics_ 13, 377–377 (2012). Article CAS Google
Scholar * Chen, X., Wen, Z., Xu, W. & Xiong, S. Granulin Exacerbates Lupus Nephritis via Enhancing Macrophage M2b Polarization. _PLoS ONE_ 8, e65542 (2013). Article ADS CAS Google
Scholar * Benson, D. D. _et al_. Gender-specific transfusion affects tumor-associated neutrophil: macrophage ratios in murine pancreatic adenocarcinoma. _J Gastrointest Surg_ 14, 1560–5
(2010). Article Google Scholar * Purandhar, K. & Seshadri, S. Age associated variations in human neutrophil and sperm functioning. _Asian Pacific Journal of Reproduction_ 2, 201–208
(2013). Article Google Scholar * Kang, Y. H. _et al_. ESM-1 regulates cell growth and metastatic process through activation of NF-kappaB in colorectal cancer. _Cell Signal_ 24, 1940–9
(2012). Article CAS Google Scholar * de Graaf, N. _et al_. PA28 and the proteasome immunosubunits play a central and independent role in the production of MHC class I-binding peptides _in
vivo_. _Eur J Immunol_ 41, 926–35 (2011). Article ADS Google Scholar * Davalieva, K. _et al_. Proteomics analysis of urine reveals acute phase response proteins as candidate diagnostic
biomarkers for prostate cancer. _Proteome Sci_ 13, 2 (2015). Article CAS Google Scholar * Shirakami, Y., Sakai, H. & Shimizu, M. Retinoid roles in blocking hepatocellular carcinoma.
_Hepatobiliary Surgery and Nutrition_ 4, 222–228 (2015). PubMed PubMed Central Google Scholar * Ma, J., Malladi, S. & Beck, A. H. Systematic Analysis of Sex-Linked Molecular
Alterations and Therapies in Cancer. _Sci Rep_ 6, 19119 (2016). Article ADS CAS Google Scholar * Kolaczkowska, E. & Kubes, P. Neutrophil recruitment and function in health and
inflammation. _Nat Rev Immunol_ 13, 159–175 (2013). Article CAS Google Scholar * Smoak, K. _et al_. Effects of Liver X Receptor Agonist Treatment on Pulmonary Inflammation and Host
Defense. _Journal of immunology (Baltimore, Md. : 1950)_ 180, 3305–3312 (2008). Article CAS Google Scholar * Mitroulis, I., Kambas, K., Anyfanti, P., Doumas, M. & Ritis, K. The
multivalent activity of the tissue factor-thrombin pathway in thrombotic and non-thrombotic disorders as a target for therapeutic intervention. _Expert Opin Ther Targets_ 15, 75–89 (2011).
Article CAS Google Scholar * Ohshiro, K. _et al_. Thrombin stimulation of inflammatory breast cancer cells leads to aggressiveness via the EGFR-PAR1-Pak1 pathway. _Int J Biol Markers_ 27,
e305–13 (2012). Article CAS Google Scholar * Yang, L. & Xu, L. GPR56 in cancer progression: current status and future perspective. _Future Oncol_ 8, 431–40 (2012). Article CAS
Google Scholar * Xu, L. _et al_. GPR56 plays varying roles in endogenous cancer progression. _Clin Exp Metastasis_ 27, 241–9 (2010). Article CAS Google Scholar * Wang, Z. _et al_.
RARRES3 suppressed metastasis through suppression of MTDH to regulate epithelial-mesenchymal transition in colorectal cancer. _American Journal of Cancer Research_ 5, 1988–1999 (2015). CAS
PubMed PubMed Central Google Scholar * Casanova, B. _et al_. The class II tumor-suppressor gene RARRES3 is expressed in B cell lymphocytic leukemias and down-regulated with disease
progression. _Leukemia_ 15, 1521–6 (2001). Article CAS Google Scholar * Xu, Y. _et al_. The antitumor effect of TIG3 in liver cancer cells is involved in ERK1/2 inhibition. _Tumour Biol_
37, 11311–20 (2016). Article CAS Google Scholar * Zhou, D. _et al_. Exome Capture Sequencing of Adenoma Reveals Genetic Alterations in Multiple Cellular Pathways at the Early Stage of
Colorectal Tumorigenesis. _PLOS ONE_ 8, e53310 (2013). Article ADS CAS Google Scholar * Gruver-Yates, A. L. & Cidlowski, J. A. Tissue-Specific Actions of Glucocorticoids on
Apoptosis: A Double-Edged Sword. _Cells_ 2, 202–223 (2013). Article CAS Google Scholar * Volden, P. A. & Conzen, S. D. The influence of glucocorticoid signaling on tumor progression.
_Brain, behavior, and immunity_ 30, S26–S31 (2013). Article CAS Google Scholar * Wang, M. _et al_. Impaired anti-inflammatory action of glucocorticoid in neutrophil from patients with
steroid-resistant asthma. _Respiratory Research_ 17, 153 (2016). Article ADS Google Scholar * Fridlender, Z. G. _et al_. Transcriptomic analysis comparing tumor-associated neutrophils
with granulocytic myeloid-derived suppressor cells and normal neutrophils. _PLoS One_ 7, e31524 (2012). Article ADS CAS Google Scholar * De Palma, M. & Lewis, C. E. Macrophage
regulation of tumor responses to anticancer therapies. _Cancer Cell_ 23, 277–86 (2013). Article Google Scholar * Tazzyman, S., Niaz, H. & Murdoch, C. Neutrophil-mediated tumour
angiogenesis: subversion of immune responses to promote tumour growth. _Semin Cancer Biol_ 23, 149–58 (2013). Article CAS Google Scholar * Mas, V. R. _et al_. Differentially Expressed
Genes between Early and Advanced Hepatocellular Carcinoma (HCC) as a Potential Tool for Selecting Liver Transplant Recipients. _Molecular Medicine_ 12, 97–104 (2006). Article CAS Google
Scholar * Stampolidis, P., Ullrich, A. & Iacobelli, S. LGALS3BP, lectin galactoside-binding soluble 3 binding protein, promotes oncogenic cellular events impeded by antibody
intervention. _Oncogene_ 34, 39–52 (2015). Article CAS Google Scholar * Liongue, C., Hall, C. J., O’Connell, B. A., Crosier, P. & Ward, A. C. Zebrafish granulocyte colony-stimulating
factor receptor signaling promotes myelopoiesis and myeloid cell migration. _Blood_ 113, 2535–46 (2009). Article CAS Google Scholar * Yan, C., Yang, Q. & Gong, Z. Tumor-Associated
Neutrophils and Macrophages Promote Gender Disparity in Hepatocellular Carcinoma in Zebrafish. _Cancer Res_ 77, 1395–1407 (2017). Article CAS Google Scholar * Zhao, Y., Huang, X., Ding,
T. W. & Gong, Z. Enhanced angiogenesis, hypoxia and neutrophil recruitment during Myc-induced liver tumorigenesis in zebrafish. _Sci Rep_ 6, 31952 (2016). Article ADS CAS Google
Scholar * Kuang, D. M. _et al_. Peritumoral neutrophils link inflammatory response to disease progression by fostering angiogenesis in hepatocellular carcinoma. _J Hepatol_ 54, 948–55
(2011). Article CAS Google Scholar * Hall, C., Flores, M., Storm, T., Crosier, K. & Crosier, P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic
fish. _BMC Developmental Biology_ 7, 42 (2007). Article Google Scholar * Lawson, N. D. & Weinstein, B. M. _In vivo_ imaging of embryonic vascular development using transgenic
zebrafish. _Dev Biol_ 248, 307–18 (2002). Article CAS Google Scholar * Martin, M. _et al_. PP2A regulatory subunit Balpha controls endothelial contractility and vessel lumen integrity via
regulation of HDAC7. _Embo j_ 32, 2491–503 (2013). Article CAS Google Scholar * Korzh, S. _et al_. Requirement of vasculogenesis and blood circulation in late stages of liver growth in
zebrafish. _BMC Dev Biol_ 8, 84 (2008). Article Google Scholar * Manoli, M. & Driever, W. Fluorescence-activated cell sorting (FACS) of fluorescently tagged cells from zebrafish larvae
for RNA isolation. _Cold Spring Harb Protoc_ 2012 (2012). * Zhao, Y., Huang, X., Ding, T. W. & Gong, Z. Enhanced angiogenesis, hypoxia and neutrophil recruitment during Myc-induced
liver tumorigenesis in zebrafish. 6, 31952 (2016). * Lam, S. H. _et al_. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. _Nat
Biotechnol_ 24, 73–5 (2006). Article CAS Google Scholar * Anders, S., Pyl, P. T. & Huber, W. HTSeq – A Python framework to work with high-throughput sequencing data (2014). * Anders,
S. _et al_. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. _Nat Protoc_ 8, 1765–86 (2013). Article Google Scholar * Chaussabel, D. _et al_. A
modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. _Immunity_ 29, 150–64 (2008). Article CAS Google Scholar * Subramanian, A. _et al_.
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. _Proceedings of the National Academy of Sciences_ 102, 15545–15550 (2005). Article
ADS CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by grants from Ministry of Education of Singapore (8). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS *
Department of Biological Sciences, National University of Singapore, Singapore, Singapore Xiaojing Huo, Hankun Li, Chuan Yan, Ira Agrawal & Zhiyuan Gong * Genome Institute of Singapore,
Singapore, Singapore Zhen Li, Sinnakaruppan Mathavan & Jianjun Liu Authors * Xiaojing Huo View author publications You can also search for this author inPubMed Google Scholar * Hankun Li
View author publications You can also search for this author inPubMed Google Scholar * Zhen Li View author publications You can also search for this author inPubMed Google Scholar * Chuan
Yan View author publications You can also search for this author inPubMed Google Scholar * Ira Agrawal View author publications You can also search for this author inPubMed Google Scholar *
Sinnakaruppan Mathavan View author publications You can also search for this author inPubMed Google Scholar * Jianjun Liu View author publications You can also search for this author
inPubMed Google Scholar * Zhiyuan Gong View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS X.H., C.Y. and Z.G. conceived experiments. X.H.,
H.L. and C.Y. performed all experiments. Z.L., I.A., M.S. and J.L. provided materials and/or analysis tools. X.H. and Z.G. analyzed data and wrote the manuscript. CORRESPONDING AUTHOR
Correspondence to Zhiyuan Gong. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains neutral
with regard to jurisdictional claims in published maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY FIGURES AND TABLES RIGHTS AND PERMISSIONS OPEN ACCESS
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as
long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third
party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the
article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright
holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Huo, X., Li, H., Li, Z. _et al._
Transcriptomic profiles of tumor-associated neutrophils reveal prominent roles in enhancing angiogenesis in liver tumorigenesis in zebrafish. _Sci Rep_ 9, 1509 (2019).
https://doi.org/10.1038/s41598-018-36605-8 Download citation * Received: 13 November 2017 * Accepted: 31 October 2018 * Published: 06 February 2019 * DOI:
https://doi.org/10.1038/s41598-018-36605-8 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
