A novel scale based on biomarkers associated with covid-19 severity can predict the need for hospitalization and intensive care, as well as enhanced probabilities for mortality
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Prognostic scales may help to optimize the use of hospital resources, which may be of prime interest in the context of a fast spreading pandemics. Nonetheless, such tools are
underdeveloped in the context of COVID-19. In the present article we asked whether accurate prognostic scales could be developed to optimize the use of hospital resources. We retrospectively
studied 467 files of hospitalized patients after COVID-19. The odds ratios for 16 different biomarkers were calculated, those that were significantly associated were screened by a Pearson’s
correlation, and such index was used to establish the mathematical function for each marker. The scales to predict the need for hospitalization, intensive-care requirement and mortality had
enhanced sensitivities (0.91 CI 0.87–0.94; 0.96 CI 0.94–0.98; 0.96 CI 0.94–0.98; all with _p_ < 0.0001) and specificities (0.74 CI 0.62–0.83; 0.92 CI 0.87–0.96 and 0.91 CI 0.86–0.94; all
with _p_ < 0.0001). Interestingly, when a different population was assayed, these parameters did not change considerably. These results show a novel approach to establish the
mathematical function of a marker in the development of highly sensitive prognostic tools, which in this case, may aid in the optimization of hospital resources. An online version of the
three algorithms can be found at: http://benepachuca.no-ip.org/covid/index.php SIMILAR CONTENT BEING VIEWED BY OTHERS DEVELOPING AND VALIDATING COVID-19 ADVERSE OUTCOME RISK PREDICTION
MODELS FROM A BI-NATIONAL EUROPEAN COHORT OF 5594 PATIENTS Article Open access 05 February 2021 DEVELOPING A COVID-19 MORTALITY RISK PREDICTION MODEL WHEN INDIVIDUAL-LEVEL DATA ARE NOT
AVAILABLE Article Open access 07 September 2020 EARLY OUTCOME DETECTION FOR COVID-19 PATIENTS Article Open access 16 September 2021 INTRODUCTION Two years and ten months after the SARS-CoV-2
pandemics started, more than 621 million cases and more than 6.54 million COVID-19-related deaths have been documented by the World Health Organization (WHO)1. In such a rapid growing
outbreak hospital resiliency has been repeatedly challenged worldwide2, imposing an important toll on the physical and mental health of healthcare workers3,4, and leading to the saturation
of regular hospital beds as well as intensive care unit (ICU) beds, thus producing a reduction on the quality of medical attention5, which may have impacted in an enhanced mortality, not
only in SARS-CoV-2 patients, but on the totality of cases. Moreover, in some developing countries healthcare centers reached a complete saturation by COVID-19 patients, leaving many
SARS-CoV-2 positive and negative patients without a hospital bed6,7. Arguably, many of the coronavirus-related hospitalizations were unnecessary, and those patients could have been treated
at home, freeing hospital resources for patients with enhanced needs. Nonetheless, a precise system to discriminate such cases in order to optimize hospital resource use is lacking. On the
other hand, while more than 80% of COVID-19 patients have a mild to asymptomatic disease, 20% of the patients present moderate to critical forms of the illness8, and mortality ranges from
less than 1–5.4%. While some drugs may prevent the progression to critical illness and mortality, their use should be timely9,10, therefore the development of a prognostic tool with enhanced
ease of use, speed and accuracy may be of paramount importance to prevent such outcomes. In this scenario, a series of prognostic scales have been developed to predict either the potential
for mortality or for aggravating disease, but they are not accurate11,12, are published in non-peer-reviewed journals13, are developed after small samples14, are not COVID-19-specific15 or
only predict mortality16; therefore many clinical practitioners may had the impression that these prognostics could be done easily and accurately, perhaps leading to the aggravation of the
problem. We think that in order to enhance the capacity of healthcare workers to optimize the use of hospital resources, including regular and ICU beds, or the administration of life-saving
drugs that are either expensive and/or scarce, three prognostic tools should be developed: one to predict the need for hospitalization, another to predict the need for intensive care (IC),
and lastly one to predict the potential for mortality. In the present article we show the development of a prognostic algorithm that is able to predict these outcomes specifically in
COVID-19 patients. The tool was developed after the study of 422 patients and shows an enhanced sensitivity (over 90%). Moreover, we made a confirmatory study with the patients of a
different hospital, finding that the sensitivity did not change considerably. An online, easy to use, version of this tool can be found on: http://benepachuca.no-ip.org/covid/index.php
Finally, the methodological approach to establish the mathematical functions of the different biomarkers by the means of the Pearson’s correlation index is, to our knowledge, novel. And we
think that may aid in the development of similar prognostic tools. MATERIALS AND METHODS We gathered a total of 467 clinical files belonging to patients that were hospitalized at Sociedad
Española de Beneficencia and Hospital Español, from March 12th 2020 to August 1st 2022 with the diagnosis of COVID-19. The records were screened for the following inclusion criteria: (i)
patients with confirmed SARS-CoV-2-derived infection (positive PCR, rapid antigen, IgM and/or CORADS 4 or higher), (ii) files that showed laboratory evaluations in the first 24 h after
hospital admission, (iii) patients with signed informed consent for the study. Then the records that complied with one or more of the following exclusion criteria were eliminated: (i)
voluntarily-discharged patients, (ii) patients with clinical files lacking crucial information, (iii) post-COVID-19 care, and (iv) patients that were referred from other institutions and
therefore had incomplete records. Relevant data about markers that were previously associated with severe COVID-19 were collected on a Google Docs file, sorting the patients according to
three different outcomes: mortality, intensive care requirement (ICR) and hospitalization requirement (HR). Mortality was defined as death occurring within hospitalization; ICR was defined
as the use of the ICU staff and facilities for at least one day; and HR was defined as patients requiring a respiratory support superior to 5 L/min for at least one day (because this
requirement is most likely unsustainable in an _at home_ treatment setting). The control patients for HR, ICR or mortality were those that did not required more than 10L/min of oxygen
supplementation, intensive care or did not died within the hospital, respectively. The percentage of lung infiltration was evaluated as described elsewhere17,18,19 using the Chest CT Score.
Briefly, chest computed tomography studies were evaluated by two independent researchers whom divided the lungs into five anatomical regions (one for each lobule), and assigned each one up
to five points depending on the percentage of the parenchyma that was infiltrated, adding the points at the end to a maximum of 25 points. The level of association of each marker with the
three different outcomes was initially assessed by calculating the odds ratio (OR), and the markers were considered to have a significant association with a particular outcome when _p_ ≤
0.05. For this test the reference values were those published elsewhere20. We then performed a Kolmogorov–Smirnov test in order to determine the type of distribution of the data (data not
shown). Then, the patients’ values were transformed in a binary manner, considering their value as “0” when they did not surpassed their reference values, and as “1” when they did. The
binary data for significant associated markers was used to perform a Pearson’s correlation to estimate their weight or mathematical function, but only those that had a Pearson’s correlation
index ≥ 0.20 were integrated into the pertinent algorithm. These consisted on the addition of the function of each marker (Pearson’s index) when the patient exceeded the marker’s reference
values. Importantly, being that our data followed a non-Gaussian distribution, we opted for the Sperman’s correction for the Pearson’s test. The individual patient’s outcome predictor (OP)
values were calculated and plotted into a Receiver Operating Characteristic (ROC) curve to estimate the sensitivity of each algorithm in the prediction of the aforementioned outcomes.
Furthermore, the mean and standard deviation of the control group (negative for each outcome) were added, while the standard deviation was deducted from the mean of the outcome-positive
group, and the middle point between each operation’s results was found to calculate the cutoff value. Sensitivity (SE), specificity (SP), positive (PPV) and negative predictive (NPV) values,
as well as the OR and Chi2 values were then calculated to investigate each algorithm’s characteristics. All the statistic tests were performed and graphed using GraphPad Prism X9, and
significant differences were considered when _p_ ≤ 0.05. A protocol for this study was evaluated by the Institutional Committee of Research Ethics of the Sociedad Española de Beneficencia
(Pachuca, Hidalgo) and the study was approved on February 24th of 2020. Our sponsor had no role in study design. All methods were performed in accordance with the relevant guidelines and
regulations, including the Declaration of Helsinki, and written informed consent was obtained from all the patients studied. RESULTS We assessed a total of 467 clinical files belonging to
patients that were hospitalized at Sociedad Española de Beneficencia and Hospital Español, from March 12th 2020 to August 1st 2022. All files were analyzed, and 422 were found to be suitable
for analysis. 255 files were allocated to algorithm design and 167 were used to validate the algorithms (Supp. Fig. S1). The only criteria for such allocation was to use the files belonging
to Hospital Español in the design of the algorithms, while the files from Sociedad Española de Beneficencia were used to validate the tools with a different population. The patients whom
contributed with the data for algorithm design were unvaccinated against SARS-CoV-2, while only 26% of the patients that provided data for the algorithms’ validation had already received
such treatment. Of the 255 clinical files that we used to calculate the algorithm 175 (74.5%) belonged to patients that were retrospectively found to have a justifiable hospitalization,
while 59 (25.5%) did not develop characteristics that made hospitalization mandatory over their whole hospital stay. Moreover, 125 (49.6%) patients required IC and 79 (31.1%) died at the
hospital. On the other hand, given that severely affected chest tomography findings (% inf)21, C-reactive protein (CRP), d-dimer, neutrophils, lymphocytes, lactate dehydrogenase (LDH)22,
procalcitonin, medium arterial pressure (MAP), creatinine, leukocytes, aspartate aminotransferase (AST)23,24, ferritin, oxygen saturation (sO2)25, and advanced age and comorbidities26 have
been associated with COVID-19 progression, the exact values of these markers were extracted from the complete clinical files of the participants. Upon gathering the laboratory data for the
first 24 h of hospitalization, the OR for each of these markers was calculated in relation to HR (Supp. Table S1), ICR (Supp. Table S2) or enhanced probability for mortality (Supp. Table
S3). We found six significant associated markers with HR, which were a Kirby index < 300 (OR 5.58, CI 1.95–15.97, _p_ = 0.0010) and Kirby index < 200 (OR 34.16, CI 15.10–70.99, _p_ =
0.0001), as well as sO2 < 90% (OR 2.15, CI 1.138–3.30, _p_ = 0.0133), sO2 < 80% (OR 3.04, CI 1.415–6.833, _p_ = 0.0043), CRP > 120 mg/dL (OR 2.95, CI 1.439–5.838, _p_ = 0.0021) and
LDH > 400 U/L (OR 2.6, CI 1.393–4.982, _p_ = 0.0034). Moreover, we found 13 significant association with ICR, which were d-dimer > 500 ng/mL (OR 2.411, CI 1.160–5.017, _p_ = 0.0179),
neutrophils > 7700 cells/µL (OR 1.807, CI 1.065–3.152, _p_ = 0.0323), % inf > 15/25 (OR 3.904, CI 1.598–9.573, _p_ = 0.0024), age > 60 years (OR 4.075, CI 1.791–8.559, _p_ =
0.0004), Kirby index < 300 (OR 23.16, CI 3.625–247.3, _p_ = < 0.0001), Kirby index < 200 (OR 11.76, CI 4.691–28.26, _p_ = < 0.0001), sO2 < 90% (OR 2.619, CI 1.347–5.031, _p_
= 0.0042), sO2 < 80% (OR 9.048, CI 3.746–21.41, _p_ = < 0.0001), CRP > 120 mg/dL (OR 2.611, CI 1.467–4.591, _p_ = 0.0008), ferritin > 150 mg/dL (OR 91.38, CI 22.49–387.3, _p_ =
< 0.0001), LDH > 211 U/L (OR 5.876, CI 1.811–19.26, _p_ = 0.0021), LDH > 400 U/L (OR 2.730, CI 1.575–4.673, _p_ = 0.0003), and AST > 70 U/L (OR 2.349, CI 1.060–5.056, _p_ =
0.0330), And finally, we found 15 markers associated with enhanced probability for mortality, which were creatinine > 1 mg/dL (OR 2.4868, CI 1.3698–4.5148, _p_ = 0.0023), d-Dimer > 500
ng/mL (OR 3.272, CI 1.271–7.970, _p_ = 0.0130), neutrophils > 7700 cells/µL (OR 2.645, CI 1.447–4.997, _p_ = 0.0018), % inf > 15/25 (OR 80.35, CI 31.95–194.4, _p_ = < 0.0001), age
> 60 years (OR 8, CI 2.22–28.9, _p_ = 0.0191), MAP < 65 (OR 4.085, CI 1.016–18.04, _p_ = 0.0451), Kirby index < 300 (OR 7.282, CI 1.141–78.42, _p_ = 0.0284), Kirby index < 140
(OR 6.517, CI 2.361–16.10, _p_ = < 0.0001), sO2 < 90% (OR 0.3199, CI 16.24–0.6408, _p_ = 0.0011), sO2 < 80% (OR 2.11, CI 1.168–3.951, _p_ = 0.0143), CRP > 120 mg/dL (OR 4.546,
CI 2.596–8.178, _p_ = < 0.0001), ferritin > 150 ng/mL (OR 8.571, CI 0.4919–147.4732, _p_ = 0.0463), > 1 comorbidity (OR 2, CI 1.0824–3.9972, _p_ = 0.0266), LDH > 211 U/L (OR
4.0854, CI 0.9290–17.9664, _p_ = 0.0451) and LDH > 400 U/L (OR 3.338, CI 1.808–5.978, _p_ = < 0.0001). The markers with significant associations were plotted into a heat map and their
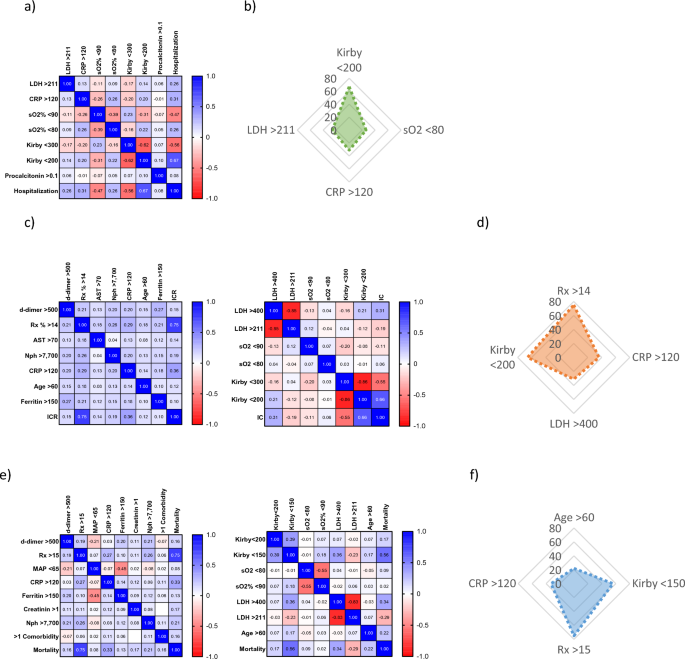
Pearson’s correlation coefficient was calculated (Fig. 1a,c,e) and used to determine the relative weight, or mathematical function, of each variable into each of the three algorithms. Only
four variables had a Pearson’s correlation index ≥ 0.20 in relation to each outcome, and thus were considered for the development of the algorithms, being Kirby < 200, LDH > 211, CRP
> 120 and sO2 < 80 important for the prognostic of HR; Rx > 14, Kirby < 200, CRP > 120, and LDH > 400 for the prediction of ICR; as well as age > 60, Kirby < 150, CRP
> 120 and Rx > 15 for mortality (Fig. 1b,d,f). To calculate the OP score for each patient, the Pearson’s index belonging to each variable (Fig. 1b,d,f) was added each time a
particular patient presented an abnormal level of a particular marker, and then both control (outcome negative) and experimental (outcome positive) patients’ values were used to calculate
the area under the ROC curve (AUROC). The COVID-hospitalization outcome prognostic (COVID-HOP) scale had an AUROC of 91% (CI 0.8725–0.9482 at 95%, _p_ < 0.0001), and both the
COVID-intensive care outcome prognostic (COVID-ICOP) and the COVID-mortality outcome prognostic (COVID-MOP) scales had an AUROC of 96% (CI 0.9448–0.9855 at 95%, _p_ < 0.0001 and CI
0.9464–0.9872 at 95%, _p_ < 0.0001, respectively) (Fig. 2). The cutoff value for the COVID-HOP scale was found to be 52.7, while the COVID-ICOP was 113.1 and the COVID-MOP was 109 (Supp.
Fig. S2). Thus, the complete algorithms with cutoff values were designed as detailed in Table 1, where the mathematical function of each marker, given by the Pearson’s correlation index,
would add each time the patient presents levels that exceed the reference values of said marker, and if the scale’s cut-off value is exceeded by such sum, the patient would be considered at
risk of either dying, needing regular hospitalization or intensive care. Furthermore, the SE, SP, PPV, NPV (Table 2) and OR (Supp. Fig. S3) for each OP scale with the use of the respective
cutoff values were calculated, finding that the COVID-HOP had a SE of 86%, SP of 74%, PPV of 90%, NPV of 94% and OR of 18.4 with a CI at 95% of 8.6–36, _p_ ≤ 0.0001. On the other hand the
COVID-ICOP had a SE of 87%, SP of 92%, PPV of 92%, NPV of 88% and OR of 88.5 with a CI at 95% of 35–191, _p_ ≤ 0.0001. Finally, the COVID-MOP had a SE of 92%, SP of 91%, PPV of 82%, NPV of
96% and OR of 131 with a CI at 95% of 47–341, _p_ ≤ 0.0001. Furthermore, 167 patients’ records belonging to a different health center and that were not used to calculate the algorithms, were
retrospectively studied to perform a validation of the SE, SP, PPV and NPV. Only 26% of these patients (32 individuals) were vaccinated against the coronavirus. The results for the MOP
algorithm showed no variation in the second population tested, while the ICOP scale exhibited only minimal variation. In regards to the HOP algorithm, the specificity was considerably
reduced (0.74 in the creation of the algorithm, 0.36 in the test of accuracy), but the other parameters remained without significant changes (Table 3). Finally, an online version of the
algorithms was developed to facilitate its use, and can be found at: http://benepachuca.no-ip.org/covid/index.php DISCUSSION In the present research we assessed the degree of correlation of
16 biomarkers (three of them with 2 different reference limits) with three different outcomes (the future need for hospitalization and/or intensive care as well as the enhanced probability
of mortality) by calculating the odds ratio, revealing six markers associated with the first outcome, 13 with the second, and 15 with the last. Nonetheless, when the data was binary
transformed and analyzed by the means of a Pearson’s correlation, only four markers were found to be associated with each marker: (i) Kirby < 200, LDH > 211, sO2 < 80 and CRP >
120 were highly associated with the requirement for hospitalization; Rx > 14, Kirby < 200, CRP > 120, and LDH > 400 were strongly related to the requirement for intensive care;
and finally, age > 60, CRP > 120, Rx > 15 and Kirby < 150 correlated with a high mortality. We then developed three different algorithms, all of them based on adding the
Pearson’s correlation index for the markers that were relevant to each outcome, every time a patient developed pathological levels of a particular molecule. Interestingly, when calculating
the mathematical functions in biomedical sciences a common approach is to perform a nomogram27, because of the underlying convenience of such technique. Nonetheless, the precision of such a
graphic tool is not remarkable. In these circumstances, the addition of the Pearson’s coefficients helped to develop a series of tools with enhanced sensitivity, as the COVID-HOP,
COVID-ICOP, and COVID-MOP algorithms showed a sensitivity over 90% in each case. Currently many meta-analysis28,29,30,31,32,33 studying the risk factors and biomarkers for prediction of
COVID-19 outcomes are available, but these are primarily based on cohort studies that are only representative of the Asian population, with minimal involvement of other genetic backgrounds.
In this instance, the aforementioned studies’ results reflect an enhanced degree of similarity for all the clinical and laboratory findings. Nonetheless, when different populations are
studied the level of association of some biomarkers with the disease outcomes varies34,35, in such a way that the evaluation of prognostic markers in different populations may be of
paramount importance to enhance the sensitivity and specificity of a prognostic tool. In accordance to this line of thought, here we present results derived from the analysis of a Mexican
population, that reflect key differences in the association of prognostic markers with outcomes of enhanced pathology, in which the absence of a positive correlation between comorbidities
and the worsening of COVID-19 stands out. However, in a validation experiment we observed that the degree of SE, SP, PPV, and NPV varied only in a slight manner, despite of using data
belonging to patients from a different hospital, and with a quarter part of them having been vaccinated (a condition that was not present in the patients that provided the data for the
elaboration of the algorithm). In any way, further research is needed to confirm if such homogeneity is paralleled in an international cohort. If the present tools does not possess enhanced
precision, the development of specific algorithms for each region may be a viable option. Finally, the chest CT evaluation is made subjectively according to the physician’s appreciation,
which could impair the results of the prognostics for enhanced mortality and intensive care requirement, as this marker has an increased weight into these algorithms. Nonetheless, excellent
new technologies appear to be emerging on the field, in which such evaluation is made accurately36,37, and its widespread use may be helpful in the homologation of prognostic criteria.
Overall, these results show the development of three tools that may aid in the administration of hospital resources, including regular hospital beds, intensive care unit beds, and drugs.
Such technology may be of enhanced utility in the context of the pandemic waves, which are expected to be a common occurrence in the coming years38, especially since no vaccine formula has
been proven to produce sterilizing immunoglobulin titers39. In fact, expert committees have agreed that healthcare digital innovations are both lacking40 and necessary41 to enhance hospital
resiliency, thus making necessary the development of this kind of tools. DATA AVAILABILITY Data is available upon reasonable request to the corresponding author Alberto Navarrete Peón at
[email protected]. ABBREVIATIONS * ICU: Intensive care unit * ICR: Intensive care requirement * HR: Hospitalization requirement * OP: Outcome prognostic * ROC: Receiver operator
characteristics * SE: Sensitivity * SP: Specificity * PPV: Positive predictive value * NPV: Negative predictive value * IC: Intensive care * %inf: Extension of the inflammatory infiltrate of
the lungs measured by tomography * CRP: C-reactive protein * LDH: Lactate dehydrogenase * MAP: Medium arterial pressure * AST: Aspartate aminotransferase * sO2: Oxygen saturation * AUROC:
Area under the ROC curve * HOP: Hospitalization-outcome prognostic * ICOP: Intensive care-outcome prognostic * MOP: Mortality-outcome prognostic REFERENCES * Organization, W.H. _WHO
Coronavirus (COVID-19) Dashboard_. 2022 05/08/2022 [cited 2022 02/08/2022]; Available from: https://covid19.who.int/. * Barbash, I. J. & Kahn, J. M. Fostering hospital resilience—lessons
from COVID-19. _JAMA_ 326(8), 693–694 (2021). Article CAS PubMed Google Scholar * Castro, H.M., Prieto, M.A., & Muñoz, A.M. Prevalence of burnout in healthcare workers during the
COVID-19 pandemic and associated factors. A cross-sectional study. _Medicina (B Aires)_ 82(4), 479–486 (2022). * Al-Amer, R. _et al._ Nurses experience of caring for patients with COVID-19:
A phenomenological study. _Front. Psychiatry_ 13, 922410 (2022). Article PubMed PubMed Central Google Scholar * Castagna, F. _et al._ Hospital bed occupancy rate is an independent risk
factor for COVID-19 inpatient mortality: A pandemic epicentre cohort study. 12(2), e058171 (2022). * Sánchez, J. _Hospitales de Hidalgo comienzan a reportar saturación de camas por covid_.
2021 [cited 2022 08/08/2022]; Available from: https://www.milenio.com/ciencia-y-salud/hidalgo-hospitales-comienzan-reportar-saturacion-camas-covid. * Molina, H., & Rodríguez, I. _Al
alza, saturación de hospitales por Covid-19_. 2022 [cited 2022 08/08/2022]; Available from:
https://www.eleconomista.com.mx/politica/Al-alza-saturacion-de-hospitales-por-Covid-19-20220117-0137.html. * Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the
coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72,314 cases from the Chinese center for disease control and prevention. _JAMA_ 323(13), 1239–1242 (2020).
Article CAS PubMed Google Scholar * Durán-Méndez, A. _et al._ Tocilizumab reduces COVID-19 mortality and pathology in a dose and timing-dependent fashion: A multi-centric study. _Sci.
Rep._ 11(1), 19728 (2021). Article ADS PubMed PubMed Central Google Scholar * Rahmah, L., _et al._ Oral antiviral treatments for COVID-19: opportunities and challenges. _Pharmacol.
Rep._ 1–24 (2022). * Ji, D. _et al._ Prediction for progression risk in patients with COVID-19 pneumonia: The CALL score. _Clin. Infect. Dis._ 71(6), 1393–1399 (2020). Article CAS PubMed
Google Scholar * Grifoni, E. _et al._ The CALL score for predicting outcomes in patients with COVID-19. _Clin. Infect. Dis._ 72(1), 182–183 (2021). CAS PubMed Google Scholar * Kamran, S.
M. _et al._ CALL score and RAS score as predictive models for coronavirus disease 2019. _Cureus_ 12(11), e11368 (2020). PubMed PubMed Central Google Scholar * Guo, J. _et al._ CURB-65
may serve as a useful prognostic marker in COVID-19 patients within Wuhan, China: A retrospective cohort study. _Epidemiol. Infect._ 148, e241 (2020). Article CAS PubMed Google Scholar *
Jang, J. G. _et al._ Prognostic accuracy of the SIRS, qSOFA, and news for early detection of clinical deterioration in SARS-CoV-2 infected patients. _J. Kor. Med. Sci._ 35(25), e234 (2020).
Article CAS Google Scholar * Kim, K. M. _et al._ Rapid prediction of in-hospital mortality among adults with COVID-19 disease. _PLoS ONE_ 17(7), e0269813 (2022). Article MathSciNet CAS
PubMed PubMed Central Google Scholar * Tiryaki, Ş, Dabeşlim, H. & Aksu, Y. Chest computed tomographic findings of patients with COVID-19-related pneumonia. _Acta Radiol. Open_
10(2), 2058460121989309 (2021). PubMed PubMed Central Google Scholar * Litmanovich, D. E. _et al._ Review of chest radiograph findings of COVID-19 pneumonia and suggested reporting
language. _J. Thorac. Imaging_ 35(6), 354–360 (2020). PubMed Google Scholar * Mruk, B. _et al._ Chest computed tomography (CT) severity scales in COVID-19 disease: A validation study.
_Med. Sci. Monit._ 27, e931283 (2021). Article CAS PubMed PubMed Central Google Scholar * Eduardo Nieto-Ortega, D.A.M.-F., & Peón, A. N. Calculadora de riesgo de progresión a
hospitalización por COVID-19, enfermedad crítica y muerte. Estudio retrospectivo observacional. Soc. Esp Bene 3, 1–15 (2022). * Contreras-Grande, J. _et al._ Chest computed tomography
findings associated with severity and mortality in patients with COVID-19. _Rev. Peru Med. Exp. Salud Publica_ 38(2), 206–213 (2021). Article PubMed Google Scholar * Kermali, M. _et al._
The role of biomarkers in diagnosis of COVID-19 - A systematic review. _Life Sci_ 254, 117788 (2020). Article CAS PubMed PubMed Central Google Scholar * Izcovich, A. _et al._ Prognostic
factors for severity and mortality in patients infected with COVID-19: A systematic review. _PLoS ONE_ 15(11), e0241955 (2020). Article CAS PubMed PubMed Central Google Scholar *
Malik, P. _et al._ Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. _BMJ Evid. Based Med._ 26(3), 107–108 (2021). Article PubMed Google Scholar *
Velavan, T. P. & Meyer, C. G. Mild versus severe COVID-19: Laboratory markers. _Int. J. Infect. Dis._ 95, 304–307 (2020). Article CAS PubMed PubMed Central Google Scholar * Fu, L.
_et al._ Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: A systematic review and meta-analysis. _J. Infect._ 80(6), 656–665 (2020). Article CAS PubMed PubMed
Central Google Scholar * Park, S. Y. Nomogram: An analogue tool to deliver digital knowledge. _J. Thorac. Cardiovasc. Surg._ 155(4), 1793 (2018). Article PubMed Google Scholar * Zhang,
L. _et al._ The common risk factors for progression and mortality in COVID-19 patients: A meta-analysis. _Arch. Virol._ 166(8), 2071–2087 (2021). Article CAS PubMed PubMed Central Google
Scholar * Shi, C. _et al._ Predictors of mortality in patients with coronavirus disease 2019: A systematic review and meta-analysis. _BMC Infect. Dis._ 21(1), 663 (2021). Article CAS
PubMed PubMed Central Google Scholar * Xie, J. _et al._ Clinical characteristics, laboratory abnormalities and CT findings of COVID-19 patients and risk factors of severe disease: A
systematic review and meta-analysis. _Ann. Palliat. Med._ 10(2), 1928–1949 (2021). Article PubMed Google Scholar * Chua, P. E. Y. _et al._ Epidemiological and clinical characteristics of
non-severe and severe pediatric and adult COVID-19 patients across different geographical regions in the early phase of pandemic: A systematic review and meta-analysis of observational
studies. _J. Investig. Med._ 69(7), 1287–1296 (2021). Article PubMed PubMed Central Google Scholar * Khodeir, M. M. _et al._ Early prediction keys for COVID-19 cases progression: A
meta-analysis. _J. Infect. Public Health_ 14(5), 561–569 (2021). Article PubMed PubMed Central Google Scholar * Melo, A. K. G. _et al._ Biomarkers of cytokine storm as red flags for
severe and fatal COVID-19 cases: A living systematic review and meta-analysis. _PLoS ONE_ 16(6), e0253894 (2021). Article CAS PubMed PubMed Central Google Scholar * Parra-Bracamonte, G.
M., Lopez-Villalobos, N. & Parra-Bracamonte, F. E. Clinical characteristics and risk factors for mortality of patients with COVID-19 in a large data set from Mexico. _Ann. Epidemiol._
52, 93-98.e2 (2020). Article PubMed PubMed Central Google Scholar * Sevilla-Castillo, F. _et al._ Both chloroquine and lopinavir/ritonavir are ineffective for COVID-19 treatment and
combined worsen the pathology: A single-center experience with severely ill patients. _Biomed. Res. Int._ 2021, 8821318 (2021). Article PubMed PubMed Central Google Scholar * Su, H. _et
al._ Multilevel threshold image segmentation for COVID-19 chest radiography: A framework using horizontal and vertical multiverse optimization. _Comput. Biol. Med._ 146, 105618 (2022).
Article CAS PubMed PubMed Central Google Scholar * Qi, A. _et al._ Directional mutation and crossover boosted ant colony optimization with application to COVID-19 X-ray image
segmentation. _Comput. Biol. Med._ 148, 105810 (2022). Article CAS PubMed PubMed Central Google Scholar * Sheikh, S. & Al-Mandhari, A. COVID-19: Questionable seasonality. _J. Coll.
Phys. Surg. Pak._ 32(6), 827–828 (2022). Google Scholar * Focosi, D., Maggi, F. & Casadevall, A. Mucosal vaccines, sterilizing immunity, and the future of SARS-CoV-2 virulence.
_Viruses_ 14(2), 1 (2022). Article Google Scholar * Bertoni, V.B., et al. _Resilience, Safety and Health: Reflections About Covid-19’ Assistance_. In _Proceedings of the 21st Congress of
the International Ergonomics Association (IEA 2021)_ (Springer International Publishing, Cham, 2021). * Capolongo, S. _et al._ COVID-19 and Healthcare Facilities: A Decalogue of Design
Strategies for Resilient Hospitals. _Acta Biomed._ 91(9s), 50–60 (2020). CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS All the authors wish to thank
Sociedad Española de Beneficencia, Pachuca, Hidalgo, for providing the technical support for the realization and publication of this study. The authors also wish to thank for the scholarship
and the education that they receive from such institution. We also thank the Information Technologies team of the Hospital for programming the online version of the COVID-OP scales. FUNDING
The publication of this article was funded entirely by Sociedad Española de Beneficencia A.C. This sponsor had no role in the design of the study, interpretation of the results or the
writing of the article. AUTHOR INFORMATION Author notes * These authors contributed equally: Eduardo Nieto-Ortega, Alejandro Maldonado-del-Arenal, Lupita Escudero-Roque and Diana Ali
Macedo-Falcon. AUTHORS AND AFFILIATIONS * Sociedad Española de Beneficencia, Av. Juárez #908, Col. La Villita, CP42060, Pachuca, Hidalgo, Mexico Eduardo Nieto-Ortega, Alejandro
Maldonado-del-Arenal, Lupita Escudero-Roque, Diana Ali Macedo-Falcon, Ana Elena Escorcia-Saucedo, Adalberto León-del-Ángel, Alejandro Durán-Méndez, María José Rueda-Medécigo, Karla
García-Callejas, Sergio Hernández-Islas, Gabriel Romero-López, Daniela Pérez-Ortega, Estephany Rodríguez-Segura, Daniela Montaño‑Olmos, Jeffrey Hernández-Muñoz, Samuel Rodríguez-Peña,
Montserrat Magos, Yanira Lizeth Aco-Cuamani, Alejandro Lechuga-Martín del Campo & Alberto N. Peón * Área Académica de Medicina, Universidad Autónoma del Estado de Hidalgo, Pachuca,
Mexico Eduardo Nieto-Ortega, Alejandro Maldonado-del-Arenal, Lupita Escudero-Roque, Diana Ali Macedo-Falcon, Ana Elena Escorcia-Saucedo, Adalberto León-del-Ángel, Alejandro Durán-Méndez,
María José Rueda-Medécigo, Karla García-Callejas, Sergio Hernández-Islas, Gabriel Romero-López, Daniela Pérez-Ortega, Estephany Rodríguez-Segura & Jeffrey Hernández-Muñoz * Escuela
Superior de Apan, Universidad Autónoma del Estado de Hidalgo, Carretera Apan‑Calpulalpan s/n, Colonia, 43920, Chimalpa Tlalayote, Hgo, Mexico Ángel Raúl Hernández-Romero, Nazareth
García-Chávez, Ana Lizeth García-Otero, Analiz Mejía-Rangel, Valeria Gutiérrez-Losada, Miguel Cova-Bonilla, Alma Delia Aguilar-Arroyo, Araceli Sandoval-García, Eneyda Martínez-Francisco,
Aldo Christiaan Jardínez-Vera & Alberto N. Peón * Universidad Anáhuac, Puebla, Mexico Daniela Montaño‑Olmos & Yanira Lizeth Aco-Cuamani * Hospital Español, Blvd. Luis Donaldo Colosio
802, El Palmar, Pachuca, Mexico Jeffrey Hernández-Muñoz, Samuel Rodríguez-Peña & Alberto N. Peón * Facultad de Medicina, Universidad Nacional Autónoma de México, Av. Universidad 3004,
Copilco Universidad, Coyoacán, 04510, Mexico City, Mexico Samuel Rodríguez-Peña & Montserrat Magos * Servicio de Imagenología, Hospital Regional de Alta Especialidad de Zumpango,
Carretera Zumpango-Jilotzingo 400 Barrio de Santiago 2a Seccion, 55600, Zumpango de Ocampo, Estado de México, Mexico Blanca Azucena Vázquez-García Authors * Eduardo Nieto-Ortega View author
publications You can also search for this author inPubMed Google Scholar * Alejandro Maldonado-del-Arenal View author publications You can also search for this author inPubMed Google Scholar
* Lupita Escudero-Roque View author publications You can also search for this author inPubMed Google Scholar * Diana Ali Macedo-Falcon View author publications You can also search for this
author inPubMed Google Scholar * Ana Elena Escorcia-Saucedo View author publications You can also search for this author inPubMed Google Scholar * Adalberto León-del-Ángel View author
publications You can also search for this author inPubMed Google Scholar * Alejandro Durán-Méndez View author publications You can also search for this author inPubMed Google Scholar * María
José Rueda-Medécigo View author publications You can also search for this author inPubMed Google Scholar * Karla García-Callejas View author publications You can also search for this author
inPubMed Google Scholar * Sergio Hernández-Islas View author publications You can also search for this author inPubMed Google Scholar * Gabriel Romero-López View author publications You can
also search for this author inPubMed Google Scholar * Ángel Raúl Hernández-Romero View author publications You can also search for this author inPubMed Google Scholar * Daniela Pérez-Ortega
View author publications You can also search for this author inPubMed Google Scholar * Estephany Rodríguez-Segura View author publications You can also search for this author inPubMed
Google Scholar * Daniela Montaño‑Olmos View author publications You can also search for this author inPubMed Google Scholar * Jeffrey Hernández-Muñoz View author publications You can also
search for this author inPubMed Google Scholar * Samuel Rodríguez-Peña View author publications You can also search for this author inPubMed Google Scholar * Montserrat Magos View author
publications You can also search for this author inPubMed Google Scholar * Yanira Lizeth Aco-Cuamani View author publications You can also search for this author inPubMed Google Scholar *
Nazareth García-Chávez View author publications You can also search for this author inPubMed Google Scholar * Ana Lizeth García-Otero View author publications You can also search for this
author inPubMed Google Scholar * Analiz Mejía-Rangel View author publications You can also search for this author inPubMed Google Scholar * Valeria Gutiérrez-Losada View author publications
You can also search for this author inPubMed Google Scholar * Miguel Cova-Bonilla View author publications You can also search for this author inPubMed Google Scholar * Alma Delia
Aguilar-Arroyo View author publications You can also search for this author inPubMed Google Scholar * Araceli Sandoval-García View author publications You can also search for this author
inPubMed Google Scholar * Eneyda Martínez-Francisco View author publications You can also search for this author inPubMed Google Scholar * Blanca Azucena Vázquez-García View author
publications You can also search for this author inPubMed Google Scholar * Aldo Christiaan Jardínez-Vera View author publications You can also search for this author inPubMed Google Scholar
* Alejandro Lechuga-Martín del Campo View author publications You can also search for this author inPubMed Google Scholar * Alberto N. Peón View author publications You can also search for
this author inPubMed Google Scholar CONTRIBUTIONS E.N.O., A.M.A., L.E.R., D.A.M.F., A.E.E.S., A.L.A., A.D.M., A.L.M.C., A.C.J.V. & A.N.P. contributed to the design of the study,
performed the statistical analysis and wrote the article; K.G.C., S.H.I., G.R.L., A.R.H.R., D.P.O., S.R.S., D.M.O., J.H.M., S.R.P., M.M., Y.L.A.C., N.G.C., A.L.G.O, A.M.R., V.G.L., M.C.B.,
A.D.A.A., A.S.G., and E.M.F., acquired the data; B.A.V.G. and M.J.R.M. critically reviewed the manuscript. CORRESPONDING AUTHOR Correspondence to Alberto N. Peón. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published
maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution
4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and
the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's
Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not
permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Nieto-Ortega, E., Maldonado-del-Arenal, A., Escudero-Roque, L. _et al._ A novel
scale based on biomarkers associated with COVID-19 severity can predict the need for hospitalization and intensive care, as well as enhanced probabilities for mortality. _Sci Rep_ 13, 9064
(2023). https://doi.org/10.1038/s41598-023-30913-4 Download citation * Received: 05 December 2022 * Accepted: 03 March 2023 * Published: 04 June 2023 * DOI:
https://doi.org/10.1038/s41598-023-30913-4 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
