Characterization of human oxidoreductases involved in aldehyde odorant metabolism
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Oxidoreductases are major enzymes of xenobiotic metabolism. Consequently, they are essential in the chemoprotection of the human body. Many xenobiotic metabolism enzymes have been
shown to be involved in chemosensory tissue protection. Among them, some were additionally shown to be involved in chemosensory perception, acting in signal termination as well as in the
generation of metabolites that change the activation pattern of chemosensory receptors. Oxidoreductases, especially aldehyde dehydrogenases and aldo–keto reductases, are the first barrier
against aldehyde compounds, which include numerous odorants. Using a mass spectrometry approach, we characterized the most highly expressed members of these families in the human nasal mucus
sampled in the olfactory vicinity. Their expression was also demonstrated using immunohistochemistry in human epitheliums sampled in the olfactory vicinity. Recombinant enzymes
corresponding to three highly expressed human oxidoreductases (ALDH1A1, ALDH3A1, AKR1B10) were used to demonstrate the high enzymatic activity of these enzymes toward aldehyde odorants. The
structure‒function relationship set based on the enzymatic parameters characterization of a series of aldehyde odorant compounds was supported by the X-ray structure resolution of human
ALDH3A1 in complex with octanal. SIMILAR CONTENT BEING VIEWED BY OTHERS ELUCIDATING THE SYNERGISTIC ROLE OF HYBRID PEPTIDE FROM _BURKHOLDERIA CEPACIA_ ENZYMES IN BIODEGRADATION OF POLYCYCLIC
AROMATIC HYDROCARBONS Article Open access 12 April 2025 AMINO ACID DEPENDENT FORMALDEHYDE METABOLISM IN MAMMALS Article Open access 16 June 2020 ENDOGENOUS ZINC NANOPARTICLES IN THE RAT
OLFACTORY EPITHELIUM ARE FUNCTIONALLY SIGNIFICANT Article Open access 28 October 2020 INTRODUCTION Olfaction is the major sense that determines flavor perception when eating; it consequently
constitutes a key determinant in food intake. In accordance with this, olfactory dysfunction leads to a decrease of food enjoyment and ingestion1 or/and reduction in well-being and quality
of life sometimes leading to depression2. Olfactory sensations are based on binding of odorant molecules on olfactory receptors within the olfactory cleft3. Odorant molecules are released in
the mouth during chewing and are transported by air to the olfactory receptor via the retronasal route. These receptors are located on the surface of the olfactory cilia, which themselves
are bathed in the olfactory mucus4,5,6,7,8. Odorant molecules must therefore pass through this mucus, which contains mostly water (95%), mucopolysaccharides (2%), enzymes, glycoproteins,
antibodies and salts. Among proteins, odorant binding proteins (OBP) belong to the lipocalin family9 and are potential odorant transporters. The nasal mucus contains many other proteins10,
and recent studies have shown that among these proteins are enzymes metabolizing odorants11,12,13,14,15, which participate in olfactory peri-receptor events. These nasal proteins are
involved in the protection of cells, including olfactory neurons, against reactive molecules (aldehyde, ester, sulfur compounds, etc.) as a first barrier. As a consequence of this metabolic
activity, these enzymes can be involved in olfactory signal termination by facilitating odorant elimination. This elimination constitutes a clearance mechanism that stops the receptor signal
and prevents olfactory receptors from saturation. Additionally, it was proposed that the newly created metabolites could modify the olfactory response due to their affinity for olfactory
receptors, which can differ from the original molecules11. It was demonstrated that metabolization of some odorants in human nasal mucus/saliva resulted in the creation of new aroma
compounds affecting the activation pattern of odorant receptors12,16,17. The involved proteins are xenobiotic metabolizing enzymes (XMEs) also called odorant metabolizing enzymes (OMEs).
Evidence that some of these XMEs also act on odorants has been reported in recent years13. The first group of XMEs is phase I enzymes that functionalize odorants with chemical reactions such
as oxidation, reduction, and hydrolysis (e.g., cytochrome P450 monooxygenases, alcohol dehydrogenases, aldehyde dehydrogenases, etc.). Their function is to biotransform xenobiotics into
more polar metabolites and provide sites for conjugation reactions. The second group is phase II enzymes; these enzymes (UDP-glucuronosyl transferases, glutathione transferases, etc.) can
directly act on xenobiotics but commonly conjugate functionalized metabolites with a polar compound to increase odorant hydrophilicity and decrease their reactivity to eliminate them more
easily. Phase III proteins include membrane transports in charge of removing hydrophilic xenobiotics from the cells when the process occurs within cells. Proteomic studies have shown the
presence of phase I and phase II XMEs in human olfactory mucus and sensory cilia10,18. Other studies reported metabolizing activity of phase II glutathione transferases and UDP glucuronosyl
transferases on odorant molecules at the olfactory level19,20,21. Additionally, oxidoreduction reactions of odorant molecules after incubation in nasal mucus were reported without
identifying these enzymes12,22. Other studies have demonstrated that these metabolic reactions are enhanced by the cofactor NAD(P)H in olfactory mucus23,24. Oxidoreductases are major phase I
enzymes that are NAD(P)H-dependent and are found in many parts of the body due to their detoxification role. For instance, their activity has been demonstrated in the buccal cavity at the
salivary25,26,27 and epithelial levels27,28. At the olfactory level, they were also shown to be expressed in mouse sensory cilia18, rodent olfactory mucus and epithelium16,19 and human nasal
mucus10,18,29. The present study aims to identify enzymes potentially involved in this odorant metabolizing activity and localize them in nasal mucus and the nasal cavity as well as
demonstrate their ability to metabolize odorants. We used mass spectrometry and immunohistochemistry to identify enzymes potentially involved in odorant metabolism. Among them, three
candidates, aldehyde dehydrogenase family 1 member A1 (ALDH1A1), aldehyde dehydrogenase family 3 member A1 (ALDH3A1), and aldo–keto reductase family 1 member B10 (AKR1B10), which are already
known to detoxicate toxic substrates that we may encounter in daily life, were selected for in vitro validation of the capacity to metabolize odorants. To do this, a panel of odorants was
tested on recombinant enzymes, and the tridimensional structure of ALDH3A1 in complex with one of the best-identified odorants was solved. RESULTS IDENTIFICATION OF THE OXIDOREDUCTASES
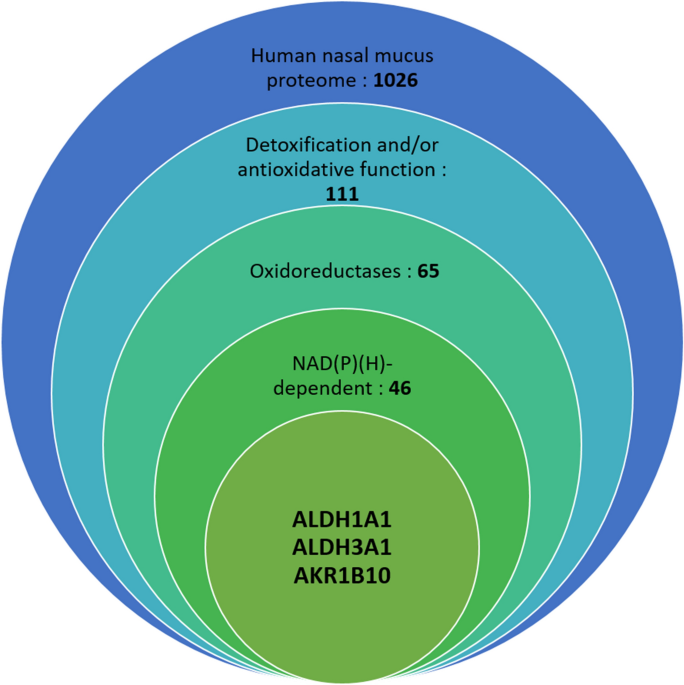
INVOLVED IN HUMAN NASAL MUCUS ODORANT METABOLISM To identify the most abundant NADH-dependent enzymes in human nasal mucus, we investigated the nasal mucus proteome from three donors by mass
spectrometry analysis, leading to the identification of 1026 different proteins. The number of identified proteins was not the same in the three donors, mainly due to the differences in the
quality of the three samples. This number is much higher than that in the proteomic analysis of Debat et al.10 in 2007, who reported 83 proteins in nasal mucus, and equivalent to that in
the analysis of Yoshikawa in 201830, in which 1236 ± 230 proteins were identified in young subjects’ mucus and 1227 ± 274 in elderly subjects’ mucus. In this study, the mucus was picked up
in the olfactory cleft using 30° rigid endoscopy. Among the most abundant proteins identified in these proteomes, we identified 111 proteins involved in detoxification or antioxidative
mechanisms or both (Fig. 1). Groups were made with UniProt classification according to previous publications, which demonstrated either a role in the detoxification of toxic compounds such
as the aldo–keto reductase family 1 member A1 (AKR1A1)31,32, a role in the antioxidant capacity by trapping or destroying free radicals such as superoxide dismutase33, or both, such as
glutathione transferase Mu 234,35,36. Additionally, enzymes and proteins already shown to be involved in human olfaction were identified in this proteome, including glutathione transferases,
GSTP120,37,38 and l-xylulose reductase (DCXR)16, and lipocalins, LCN139, LCN239, and OBPIIa14,40, also known as odorant binding proteins, as they can bind odorants (Supplemental Table 6).
Among the 1026 proteins detected, 46 NAD(P)(H)-dependent enzymes were identified and classed, as shown in Table 1. The spectra numbers indicate the total number of counts for all peptides
included and detected for the same protein. The spectra numbers are dependent on the protein size as well as the peptide stability; they are also highly driven by the protein abundance. The
two most represented NAD(P)(H)-dependent oxidoreductases in the mass spectrometry analysis based on spectral number are aldehyde dehydrogenase 1A1, also named retinal dehydrogenase 1
(ALDH1A1), and aldehyde dehydrogenase 3A1, also named dimeric NADP-preferring (ALDH3A1). They also appear at the second and third positions of the most represented proteins in terms of
spectra number among the 1026 identified proteins. Interestingly, they appear at higher level (considering the spectra number) compared to well-known mucus proteins as OBPs. In total, 11
aldehyde dehydrogenases were identified in the three human olfactory mucus samples. Aldo–keto reductases are another family of enzymes that have been identified, especially aldo–keto
reductase 1B10 (AKR1B10). It is a highly represented aldo–keto reductase among the three tested people, and this human enzyme was reported to efficiently catalyze the oxidation of toxic
aldehydes41. These three enzymes were selected to determine their role in odorant reduction and oxidation and for further immunolocalization studies on olfactory epithelium samples.
DETECTION OF ALDEHYDE DEHYDROGENASE 1A1 (ALDH1A1), ALDEHYDE DEHYDROGENASE 3A1 (ALDH3A1), AND ALDO–KETO REDUCTASE 1B10 (AKR1B10) IN HUMAN OLFACTORY EPITHELIUM AND TURBINATE During aging, the
respiratory epithelium progressively takes the place of the olfactory epithelium, including turbinates42, resulting in a mix between the olfactory and respiratory epithelium at the top of
the nasal cavity43. Consequently, it is very difficult to obtain only olfactory epithelium due to its small size (between 1 and 2 cm244) and restricted access. We selected a human epithelium
close to the cribriform plate and a sample from the inferior turbinate. In the majority of cases, the human nose includes three turbinates per side: the superior, middle and inferior
turbinates. Western blot analysis supported the expression of these three oxidoreductases (ALDH1A1, ALDH3A1, and AKR1B10) close to the olfactory epithelium, in addition to their expression
within the human nasal mucus observed by proteomic analysis. Western blots were performed using sample tissue (human olfactory vicinity epithelium and human inferior turbinate) for each
oxidoreductase and are represented in Fig. 2. The three tested oxidoreductases appear to be expressed in the cytosol of the human inferior turbinate and the human olfactory vicinity
epithelium. As a positive control, we confirmed that each antibody binds to the corresponding recombinant human enzyme (Lane 2 of Fig. 2A–C). For the three antibodies, the main bands
corresponding to the recombinant proteins ALDH1A1, ALDH3A1 and AKR1B10 appeared at the expected theoretical molecular mass of the monomer, 54.9 kDa, 52 kDa, and 36.8 kDa, respectively. The
bands corresponding to the three enzymes were observed at the expected sizes corresponding to the same sizes observed for the corresponding recombinant proteins. In some cases, additional
lower bands are observed. These bands could correspond to the degradation of the corresponding enzyme observed for ALDH3A1 in Panel B, probably due to freeze‒thaw cycles of the samples or
proteolysis activity prior to conservation. The upper band observed for recombinant ALDH1A1 could correspond to a higher oligomeric state of this enzyme preserved during SDS PAGE.
OXIDOREDUCTASES LOCALIZATION IN HUMAN OLFACTORY/RESPIRATORY EPITHELIUM To investigate the localization of the three oxidoreductases within the tested tissues of the olfactory cleft,
immunohistochemistry was performed to stain ALDH1A1, ALDH3A1, and AKR1B10 in the human olfactory vicinity and human inferior turbinate epithelium from two different human donors. Both the
turbinate and olfactory vicinity contain the three oxidoreductases, as supported by the Western blot analysis (Fig. 3). Three oxidoreductases were found in both tissues. Whereas ALDH1A1 and
ALDH3A1 showed a relatively high signal intensity, the immunohistochemical signal was much lower for AKR1B10. It appears that the three oxidoreductases are synthesized in the major
epithelial cell types, since DAB signals were verified in almost every cell in our samples. Additionally, we identified DAB staining of the three oxidoreductases on the apical, ciliated
surface of the samples. This allows for interaction with the molecules, including odorants, which come in contact with and can penetrate these cells. No staining was observed in the goblet
cells (Gc) for the three tested enzymes. The absence of the three enzymes in the goblet cells involved in mucus secretion suggests secretion by nasal glands to explain their high expression
in the human nasal mucus. In comparison to the oral cavity, ALDH3A1, which is also found in human saliva, is secreted by the salivary glands45, as is the case for other aldehyde
dehydrogenases46. AKR1B10 as ALDH1A1 and ALDH3A1 also lack the conventional signal peptide at the N terminus. However, a molecular chaperone, Hsp90α associates with AKR1B10 (toward a
α-helix), then transports it to lysosomes, and is secreted jointly with Hsp90 out of the cell47. In this context AKR1B10 can potentially be directly secreted in the mucus by the ciliated
cells. Interestingly, Hsp90α is also significantly found (numerous unique peptide and numerous count) in the mucus of the three tested donors (Supplemental Table 6). OXIDOREDUCTASES
METABOLIZE ODORANT MOLECULES To test the capacity of the three selected oxidoreductases to oxidize or reduce odorant molecules, the three enzymes were recombinantly produced in _Escherichia
coli._ Then, they were purified using chromatography columns to perform enzymatic assays. The three enzymes were obtained at a high level of purity > 98% (Supplemental Fig. 1). The three
enzymes used either nicotinamide adenine dinucleotide (NAD+, for ALDH1A1 and ALDH3A1) or nicotinamide adenine dinucleotide phosphate (NADPH, for AKR1B10) as cofactors. Consequently, the
reaction can be monitored by the reduction of NAD+ to NADH or the oxidization of NADPH to NADP, as the reduced forms absorb the light at 340 nm in contrast to the oxidized form. A panel of
twenty odorants all belonging to the aldehyde class were selected. The twenty selected odorant aldehydes are aliphatic or aromatic aldehydes (Table 2). In the presence of NAD+, the two ALDHs
can catalyze the oxidation of aldehydes into their corresponding carboxylic acids, while AKR catalyzes the reduction of aldehydes into their corresponding alcohols in the presence of NADPH.
For each enzyme presented here, a Michaelis response was observed in accordance with the few substrates previously tested in the literature48,49 and allowed for calculation of the kinetic
parameters (KM, kcat). Both tested ALDHs can metabolize aliphatic or aromatic aldehydes but not with the same efficiency. ALDH3A1 has very high efficiencies in metabolizing medium-chain
aliphatic aldehydes such as heptanal, octanal, and nonanal (129, 209, and 164 min−1 µM−1, respectively) as well as aromatic aldehydes such as hydrocinnamaldehyde (169 min−1 µM−1)
(Supplemental Fig. 2). These high efficiencies are mainly driven by high catalytic constants toward these compounds (between 4000 and 7000 min−1), whereas the Michaelis constants are higher
than those of ALDH1A1 (Supplemental Table 1). ALDH3A1 metabolizes all the aliphatic aldehydes tested to their carboxylic acid relatives, except propanal. The results also show a progressive
increase in the efficiency of ALDH3A1 from butanal up to octanal, where it reaches its maximum, before decreasing for carbon chain lengths above 8 (Table 2). This evolution in efficiency
seems to be driven by the Michaelis constant KM, which reaches its minimum for octanal and increases depending on the carbon chain length. The efficiency of ALDH1A1 varies little according
to the length of the carbon chain or the presence of an aromatic ring (Table 2), and its maximum efficiency is obtained with pentanal (30 min−1 µM−1). The kcat of this enzyme is low
(approximately 50 min−1), which strongly contributes to lower efficiencies toward the tested odorants compared to ALDH3A1. However, the measured KM was the best for ALDH1A1 compared with the
two other enzymes (Supplemental Table 1). AKR1B10 catalyzes the reduction of aldehydes to the corresponding alcohols via the oxidation of NADPH to NADP+41. AKR1B10 catalyzes the reduction
of all the aldehydes tested (Table 2) except propanal. The best efficiencies were measured for hexanal, hydrocinnamaldehyde, and vanillin (25, 25, and 45 min−1 µM−1, respectively). AKR1B10
metabolizes aliphatic and aromatic aldehydes without large differences in efficiency between the two types. To better understand the involved molecular interactions between enzymes and
odorous compounds during metabolization in the nasal cavity, ALDH3A1, which presents the best efficiency toward odorant molecules as well as good expression within the olfactory mucus and
the different epithelium tested within the olfactory vicinity, was studied by crystallography in complex with its better substrates. TRIDIMENSIONAL STRUCTURE OF THE ALDH3A1/OCTANAL COMPLEX
To determine the X-ray structure of ALDH3A1 in complex with an odorant, different aldehydes presenting good catalytic efficiency were tested: trans-2-nonenal, hydrocinnamaldehyde, and
octanal. Among the different tests, the structure of ALDH3A1-octanal was successfully solved (Fig. 4A). The ALDH3A1 crystal was soaked in its mother liquor containing 10 mM octanal. This
resulted in a homodimeric structure at 1.80 angström resolution, where each active site of ALDH3A1 is occupied by one octanal molecule (Fig. 4B). Interpretation of the electron density maps
in the active site region near the catalytic Cys 24350 led to the conclusion that octanal is present as two alternative conformations. Considering the carbon atoms’ positions, these two
conformations are very close, with hydrophobic interactions stabilizing the aliphatic moiety of octanal by the surrounding residues’ side chains (Tyr 65, Tyr 115, Asn 118, Leu 119, Ile 394).
This elongated hydrophobic pocket seems well suited for the binding of long-chain aliphatic as well as aromatic aldehydes such as those catalyzed by ALDH3A1, in accordance with our
enzymatic analysis. The two conformations of octanal only differ in the positions of the oxygen atom of the aldehyde group. The first conformation of octanal, is hydrogen bonded with Glu 209
via a water molecule. In this case, the side chain of Cys 243 is oriented toward Asn 114 in close vicinity within the active site. The second conformation of octanal is such that its oxygen
atom is hydrogen-bonded with Asn 114 via a water molecule. This residue (Asn 114) could be involved in the stabilization of the oxyanionic form of the hemithioacetal state during catalysis.
Our structure likely corresponds to the step just before, obtained because of the absence of NAD cofactor needed to complete the catalytic turnover. Thus, the side chain of Cys 243 is
oriented toward the octanal molecule, ready for nucleophilic attack. Our results support the role of Cys 243 as a catalytic residue and Asn 114 as a probable catalytically important residue
during catalysis. Taken together, our results show at the molecular level how an odorant aldehyde is metabolized in the ALDH3A1 active site, which is adapted for both aliphatic and aromatic
aldehydes. DISCUSSION Aldehyde molecules are found in numerous natural odors; additionally, they are used to enhance a range of fragrance notes. For example, octanal, nonanal, and decanal
are commonly used in the perfume industry for their green-floral fragrance51. Aldehydes are also frequently encountered in food; indeed, more than 300 food products contain aldehydes as
natural constituents or flavoring additives and aromas. Vanillin (vanilla), cinnamaldehyde (cinnamon) and octanal (grape, lemon, peel oil) are the most commonly used compounds52. Aldehyde
can also have an endogenous origin, synthesized by cells during lipid peroxidation, such as 4-hydroxynonenal52, which increases oxidative stress and was already shown to be well metabolized
by ALDH3A153,54. 4-hydroxynonenal as well as acetaldehyde, are suspected in the pathogeny of different diseases. In addition, aldehydes in a general manner can be toxic depending on their
concentration, which supports the importance of an efficient detoxification system in the most exposed area of the body. In this study, we observed high expression of xenobiotic metabolism
enzymes in the human nasal mucus sampled in the olfactory cleft of three different people. Some enzymes identified in these proteomes as GSTs or DCXR were previously shown to be involved in
human odor perception16,20,37,38. Additionally, new enzymes, potentially involved in odorant metabolization appear as interesting targets for further studies as the sulfotransferase
(SULT1A1). Moreover, numerous proteins allowing to maintain the enzyme function as heat shock protein or thioredoxin were also identified. For each person, enzymes involved in aldehyde
metabolism were found within the ten most represented proteins in terms of spectra numbers among the 1026 identified proteins. From a larger perspective, 65 oxidoreductase enzymes were
identified in the three proteomes, including enzymes involved in aldehyde metabolism as well as enzymes involved in reactive oxygen species reduction. Bathing of the neuron's cilia in
the mucus allows for the first step of odorant perception due to the interaction of odorant molecules with receptors located on the membrane of these neurons. Aldehydes are highly toxic to
neurons55 and need to be particularly protected and continuously renewed for less than one month for rodent olfactory neurons56,57. In this context, to safeguard an acute sense of smell,
metabolization of aldehyde compounds, including aldehyde odorants, appears essential within the mucus. In this study, glutathione transferase P1, already shown to be able to metabolize
aldehyde odorants such as cinnamaldehyde20 or to participate in the antioxidant system58, appears to be the most expressed glutathione transferase within the mucus. Previous proteomic
analysis of the human nasal mucus showed the presence of glutathione transferases; here, we highlight for the first time their abundance, revealing their important expression. Two aldehyde
dehydrogenases (ALDH1A1 and ALDH3A1) known to metabolize aldehyde molecules are among the ten highly expressed proteins in the three proteomes. Aldo–keto reductases, including AKR1B10, can
also metabolize aldehyde compounds and appear to be well expressed. These three enzymes are also well expressed in the different epithelia tested within the olfactory cleft, as supported by
western blot analysis and immunohistochemistry. These last experiments showed good expression of these three enzymes in the different epithelia, including the ciliated cells located at the
surface of the epithelia. Consequently, ALDH1A1, ALDH3A1, and AKR1B10 appear to be the first barrier against the toxicity of aldehyde compounds due to their location. It is not excluded that
the concentration of these enzymes can be lower in the olfactory epithelium or the mucus secreted by the Bowman glands due to a potential absence of their expression. However, it is most
likely that the olfactory epithelium consists of many spots within the respiratory epithelium, consequently the mucus composition will not be dramatically different compared to the one
presented in this study (probably a secreted mucus mixture from both tissue types). These three enzymes metabolize various aldehyde odorants with different profiles of efficiency for each
enzyme. The twenty tested aldehyde odorants were metabolized. Some were specifically metabolized by a specific enzyme, such as vanillin, which was only metabolized by AKR1B10 or propanal by
ALDH1A1, while others were metabolized by the three different enzymes. Their metabolization leads to the formation of the corresponding carboxylic acids (ALDHs) or the corresponding alcohols
(AKR), both of which are generally less reactive than their aldehyde precursors. Additionally, 9 other ALDHs and 7 other AKRs were identified in the mucus sampled in the olfactory cleft. To
date, 19 ALDHs59 and 13 AKRs60 have been identified in the human genome, and 11 ALDHs and 8 AKRs were identified in the nasal mucus of the three people tested in this study, showing that
most of the oxidoreductase enzymes belonging to these two families are expressed in this mucus. This large representation of these two enzymatic families is not surprising, as olfactory
tissues are continuously in contact with volatile organic compounds. This highlights the importance of detoxifying aldehyde odorants in this part of the body. The main evolutive driver to
preserve functional enzymes metabolizing aldehyde odorant compounds in the olfactory cleft is probably the tissue and very likely the olfactory neurons preservation. Additionally, these
oxidoreductases contribute to human olfaction. Octanal, the aldehyde molecule with the highest catalytic efficiency (among the 20 tested odorant molecules) for ALDH3A1, is more than 10 times
more metabolized than the two other tested oxidoreductases. This observation and the high expression of ALDH3A1 in the mucus and in ciliated cells support the major role of this enzyme in
octanal metabolization that was previously observed in vivo in human subject breath61. ALDH3A1 catalyzes the oxidation of octanal to octanoic acid; however, octanoic acid presents a strong
goat cheese odor, which is consequently different from the typical lemon scent characteristic of octanal. The ALDH3A1 active site is well tuned to catalyze octanal oxidation, as revealed by
the first structure in complex with a substrate described in this work. The active site configuration supports the enzymatic data obtained for the panel of tested odorant aldehydes. The
ALDH3A1 active site includes a hydrophobic entry pocket adapted for the binding of both aromatic and medium-chain aliphatic aldehydes. Their binding, near the catalytic Cys 243 previously
shown to be an essential catalytic residue62 (2.89 Å between the sulfur of the cysteine and the carbon of the aldehyde functional group), enables further catalysis to yield the corresponding
carboxylic acids. In addition to the two ALDHs and the AKR in this study, other isoforms among the 11 ALDHs and the 8 AKRs identified can potentially efficiently metabolize aldehyde
odorants, supporting a complex combinatory contribution in human olfaction. ALDH and AKR expression are regulated by dietary habits, supporting an adaptation of their activity toward
aldehyde odorants conditioned by these habits63,64. Additionally, human oral bacteria are also subject to variations in diet habits, presenting aldehyde activity65 and adding potential
players in human odorant perception, as already proposed for other oral bacterial activities66,67. Interestingly, ALDH2 found during this study in the nasal mucus shows a polymorphism
associated with sweet preference68, indicating a link of ALDH with flavor perception in a more general manner. In conclusion, this study provides new results regarding the identification of
key oxidoreductases involved in human perception in addition to a comprehensive enzymatic analysis of their aldehyde substrates. The structural information obtained in this study clearly
supports the role of these enzymes in odorant aldehyde metabolism, which both preserves olfactory tissue and modulates human olfaction. METHODS CHEMICALS All odorant compounds were purchased
from Sigma-Aldrich (St. Louis, MO, USA). The common name, CAS number, and catalog number are indicated in the supplemental Table 2. PREPARATION OF HUMAN SAMPLES Tissue from the vicinity of
the olfactory region was obtained from 45- and 64-year-old male patients undergoing endoscopic routine sinus surgery. The specimens were taken from the mucosa close to the superior turbinate
in direct vicinity to the olfactory area using a 30° rigid endoscope and atraumatic surgical forceps. The patients gave informed consent for participation, and the study was approved by the
Ethics Board of the Medical Faculty of Wuerzburg University, Germany (No. 179/17XX). Immediately after harvest, the samples were stored in physiological serum for a few minutes before each
sample was separated into two parts in the laboratory. One part was frozen in liquid nitrogen for western blot analysis, and the other was immersed in a buffered fixative solution for
immunohistochemistry experiments. The samples of human nasal mucus were taken from three healthy subjects (aged 31–63, 2 females, 1 male) from the region of the olfactory cleft of both sides
with a cotton swab under endoscopic control. The study was performed according to the guidelines of the Declaration of Helsinki and has been formally approved by the Dresden Hospital Ethics
Committee. PROTEIN ASSAY Tissues for western blots were defrosted and solubilized in 200 µL of 50 mM Tris–HCl pH 7.5, 250 mM saccharose, and 1 mM EDTA by two tissue-lyser cycles of 60 s
each. Then, they were centrifuged for 10 min at 10,000_g_ at 4 °C. The supernatants were recovered and ultracentrifuged at 105,000_g_ at 4 °C for 60 min to separate soluble cytosol from
insoluble microsomes. The microsomes were resuspended in 100 µL of 150 mM Tris–HCl buffer pH 8, and the cytosol and microsome fractions were stored at −80 °C. The protein levels of all
fractions were quantified by the Lowry method by using bovine serum albumin as a standard. PROTEOMIC DATA ANALYSIS Raw data collected during nano LC–MS/MS analyses were processed and
converted into an *.mgf peak list format with Proteome Discoverer 1.4 (Thermo Fisher Scientific). MS/MS data were analyzed using the search engine Mascot (version 2.4.0, Matrix Science,
London, UK) installed on a local server. Searches were performed with a tolerance on mass measurement of 0.2 Da for precursor and 0.2 Da for fragment ions against a composite target-decoy
database (20,506 × 2 total entries) built with a human Swissprot database (taxonomy 9606, January 2019, 20,388 entries) fused with the sequences of recombinant trypsin and a list of
classical contaminants (118 entries). Cysteine carbamidomethylation, methionine oxidation, protein N-terminal acetylation, and cysteine propionamidation were searched as variable
modifications. Up to one missed trypsin cleavage was allowed. The identification results were imported into Proline software (http://proline.profiproteomics.fr) for validation69. Peptide
spectrum matches taller than nine residues and ion scores > 10 were retained. The false discovery rate was then optimized to be below 1% at the protein level using the Mascot Modified
Mudpit score. Spectral counting analyses were performed with Proline 2.0. WESTERN BLOT ANALYSIS Thirty micrograms of protein equivalent of soluble cytosol from the olfactory vicinity
epithelium and soluble cytosol from the inferior turbinate and 0.05 µg of recombinant protein were loaded onto a 4–15% precast SDS‒PAGE gel using a Precision Plus Protein™ Dual Xtra Standard
molecular weight ladder. Protein migration was performed in tris–glycine-SDS buffer at 200 V for 45 min. The results from the gel were transferred onto a nitrocellulose membrane with a
Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, USA). The membrane was then bathed in 0.02 M Tris, 0.15 M NaCl, 0.1% (v/v) Tween 20 at pH 7.6 (TBST) and 5% (w/v) dry milk for 1 h with
agitation at room temperature. After five washes in TBST, the membrane was incubated with a dilution of primary antibodies mouse anti-ALDH1A1 (MA5-34924, Thermo Fisher Scientific, Waltham,
USA) diluted 1:5000, mouse anti-ALDH3A1 (sc-376089, Santa Cruz Biotechnology, Dallas, USA) diluted 1:1000, and rabbit anti-AKR1B10 (PA5-22036, Thermo Fisher Scientific, Waltham, USA) diluted
1:3000 overnight at 4 °C with agitation. After five washes in TBST, the membrane was incubated for 1 h at room temperature with agitation in TBST with goat anti-mouse HRP secondary antibody
(P0447, Agilent, Santa Clara, USA, 1:12,500) for both ALDH types and with the goat anti-rabbit HRP secondary antibody (P0448, Agilent, Santa Clara, USA, 1:12,500) for AKR1B10. After five
washes, the membrane was revealed by soaking for one minute in a mixture of 1.5 mL Luminol/enhancer solution and 1.5 mL Peroxide Reagent solution from the ECL clarity western substrate
Bio-RadTM pack. The membrane was then placed in a ChemiDocTM acquisition system, and images were acquired by luminescence every 6 s for 10 min and analyzed using Image LabTM 4.0.1 Software
(Bio-Rad). A full image for each gel is shown in Supplemental Fig. 3. IMMUNOHISTOCHEMISTRY Turbinate tissues were fixed with formaldehyde solution 4% buffered pH 6.9 (1.00496, Merck,
Darmstadt, Germany) for 48 h at room temperature. After decalcification with 10% ethylenediaminetetraacetic acid disodium salt in phosphate-buffered saline pH 7.4 for four weeks with regular
changes, the specimens were dehydrated and embedded in paraffin. The olfactory vicinity tissues were fixed in Roti-Histofix (4%, pH 7, Carl Roth, Germany) and embedded in paraffin using the
Microm STP 120 Spin Tissue Processor (Thermo, Waltham, USA). Five-micrometer-thick sections were deparaffinized, rehydrated, and stained immunohistochemically. An antigen pretreatment step
was carried out using high-temperature antigen unmasking techniques with target retrieval in citrate buffer pH 6.0 (S2369, Agilent, Santa Clara, USA) for 45 min. Endogenous peroxidases were
treated with blocking reagent (S2003, Agilent, Santa Clara, USA) for 10 min at room temperature prior to equilibration in 0.05 M Tris–HCl, 0.15 M NaCl, 0.05% Tween 20, pH 7.6. Tissue
sections were saturated for 45 min with 10% normal goat serum (G9023, Merck, Darmstadt, Germany) in antibody diluent (S0809, Agilent, Santa Clara, USA) to reduce nonspecific binding.
Sections were then incubated with the same primary antibodies as for the western blots overnight at 4 °C in the antibody diluent; primary antibodies included AKR1B10 diluted 1:4000, ALDH3A1
diluted 1:4000 and ALDH1A1 diluted 1:5000 and at 1:200 for olfactory marker protein (OMP). This last antibody (sc-365818, Santa-Cruz Biotechnology, Dallas, USA) is proposed to be specific
toward human olfactory chemosensory neurons.. Tissue sections were then incubated for 1 h at room temperature for ALDH1A1, ALDH3A1 and OMP experiments with the goat anti-mouse HRP secondary
antibody (used for the western blots) at 1:200 and with the goat anti-rabbit HRP secondary antibody (used also for the western blot) for AKR1B10 at 1:200. Due to the lack of specificity of
the anti-OMP antibody, we could not show any neuronal specific staining (Supplemental Fig. 5). Negative controls were prepared by replacing the primary antibody with antibody diluent alone
(Supplemental Fig. 4). Immunohistochemical staining was performed using a liquid DAB+ substrate chromogen system (K3468, Agilent, Santa Clara, USA). Sections were counterstained with Mayer’s
hemalum solution (1.09249, Merck, Darmstadt, Germany). The slides were examined with an Eclipse E600 microscope. Images were acquired with a DS-Ri2 digital camera using the software
NIS-Elements Basic Research (all from Nikon, Tokyo, Japan). PROTEIN PRODUCTION AND PURIFICATION The DNA sequences encoding _human_ ALDH1A1 (UniProt code P00352), ALDH3A1 (UniProt code
P30838), and AKR1B10 (UniProt code O60218) were optimized for expression in _E. coli,_ and the sequence GC rate was modified to approximately 50%. They were subcloned into the pET24b,
pET22b, and pET26b vectors between the _NdeI_ and _SacI_ restriction sites. A sequence encoding 6 histidines was added at the N-terminal extremity of ALDH3A1 and AKR1B10 for purification.
The bacterial strains _E. coli_ BL21(DE3) Star and BL21(DE3) pLysS with the following genotypes, F–_omp_T _hsd_SB (rB–, mB–) _gal dcm rne_131 (DE3) and pLysS F–_omp_T _hsd_SB (rB–, mB–) _gal
dcm_ (DE3) pLysE(CamR), were used to express both ALDHs and AKR1B10. The transformed cells were grown at 37 °C in LB medium (containing 100 μg mL−1 ampicillin) and induced by the addition
of isopropyl β-d-1-thiogalactopyranoside (IPTG) when the cell culture reached the selected OD measured at 600 nm. The IPTG concentration, time, and temperature of growth after induction
changes between the three proteins are summarized in Supplemental Table 3. Bacterial growth was stopped by centrifugation (4000_g_, 15 min), and bacteria were suspended in Tris buffer
containing 50 mM saccharose at 250 mM pH 8.0. Cells were sonicated at 4 °C and centrifuged at 24,000×_g_ for 45 min at 4 °C. The recombinant proteins within the supernatant were purified in
a first purification step consisting of two successive ammonium sulfate precipitations. Then, the salt was eliminated by two dialyzes in the appropriate buffer for each protein of interest.
Two chromatography steps, indicated in Supplemental Table 4, were performed for each enzyme to obtain the pure protein (Supplemental Fig. 1). Proteins were stored at −20 °C. ENZYMATIC ASSAYS
Enzymatic activity was determined on a UV-1800 spectrophotometer (Shimadzu, Japan) by measuring the absorbance at 340 nm, which corresponds to the NAD(P)H absorbance wavelength. Enzymatic
reactions were performed in a 1 mL quartz cuvette filled with 1 mL of a mixture containing 100 mM KPi buffer pH 7.0, a saturating concentration of cofactor, which was 1 mM NAD for both ALDH
and 200 µM NADPH for AKR1B10, an odorous molecule diluted in methanol, 500 nM ALDH1A1 or 70 nM ALDH3A1 or 250 nM AKR1B10, and water to volume. Each experiment was repeated two times. The
initial velocities for an increasing range of odorant concentrations were measured, and the Michaelis‒Menten curve was plotted using SigmaPlot software according to the equation vi = (Vmax ×
[S])/(KM + [S]), where vi is the initial rate in µM min−1, Vmax is the maximum initial rate in µM min−1, [S] is the substrate concentration in mol L−1, and KM is the Michaelis constant in
µM. The catalytic constant kcat was obtained by dividing Vmax by the enzyme concentration. The efficiency (kcat/KM) was obtained by dividing kcat by their corresponding KM. Standard errors
(Δ) of the efficiency were calculated using the equation: \(\Delta efficiency = efficiency \times \sqrt{\left({\left(\frac{\Delta {k}_{cat}}{{k}_{cat}}\right)}^{2}+{\left(\frac{\Delta
{K}_{M}}{{K}_{M}}\right)}^{2}\right)}.\) The averages of each kinetic parameter and standard error were calculated and are summarized in Supplemental Table 1. When the enzymatic activity did
not follow a Michaelis-response (due to a high KM value), the absorbance increased linearly with substrate concentrations, making the calculation of the KM value impossible; this is
represented by “nm” for “not measurable” in Table 2 and Supplemental Table 1. CRYSTALLIZATION AND X-RAY DIFFRACTION EXPERIMENTS Crystallogenesis tests were undertaken with enzymes that had a
purity level greater than 98% according to an estimate by SDS‒PAGE gel (Supplemental Fig. 1). Before the crystallization assays, ALDH3A1 was dialyzed against 10 mM pH 7.8 HEPES buffer.
Crystallization trials were performed manually at 20 °C by using the sitting drop vapor diffusion method. ALDH3A1 (2 mg ml−1) was crystallized by mixing 1 µL of protein with 1 µL of a
solution containing 18% PEG 3350 in 0.1 M potassium acetate pH 7.5 buffer. To obtain complexes of ALDH3A1 with octanal, crystals were soaked into the mother liquor plus 10 mM octanal.
Cryoprotection was achieved by adding 20% glycerol to the drops containing the crystals. The crystals were flash-frozen in liquid nitrogen before synchrotron data collection. Diffraction
experiments were performed on the SOLEIL synchrotron beamline PROXIMA1. Crystals of ALDH3A1-octanal diffracted to 1.80 Å. The datasets were indexed and integrated with XDS70 and scaled with
pointless71. The structure was solved by molecular replacement using the coordinates of the unbound form of ALDH3A1 (PDB code 3SZA). The 3D structure was manually adjusted with COOT72 and
refined with PHENIX73. Inspection of the electron density maps around the active site region allowed for the identification and building of ligands. Restraint files for ligand refinement
were generated with the GRADE webserver (http://grade.globalphasing.org). The structure was validated with MolProbity74. The figure was prepared using PyMOL (The PyMOL Molecular Graphics
System, Version 2.0 Schrödinger, LLC). The coordinates, structure factors, and diffraction statistics (Supplemental Table 5) have been deposited in the Protein Data Bank under accession
codes 8BB8 (ALDH3A1-octanal). DATA AVAILABILITY All data generated or analysed during this study are included in this published article and its supplementary information files. The crystal
structure of the ALDH3A1-octanal complex is accessible under the PDB code 8BB8 (https://www.rcsb.org/). REFERENCES * Mainland, J. D. _et al._ Identifying treatments for taste and smell
disorders: Gaps and opportunities. _Chem. Senses_ 45, 493–502. https://doi.org/10.1093/chemse/bjaa038 (2020). Article CAS PubMed PubMed Central Google Scholar * Schafer, L., Schriever,
V. A. & Croy, I. Human olfactory dysfunction: Causes and consequences. _Cell Tissue Res._ 383, 569–579. https://doi.org/10.1007/s00441-020-03381-9 (2021). Article PubMed PubMed Central
Google Scholar * Beauchamp, J., Scheibe, M., Hummel, T. & Buettner, A. Intranasal odorant concentrations in relation to sniff behavior. _Chem. Biodivers._ 11, 619–638.
https://doi.org/10.1002/cbdv.201300320 (2014). Article CAS PubMed Google Scholar * Buck, L. & Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for
odor recognition. _Cell_ 65, 175–187. https://doi.org/10.1016/0092-8674(91)90418-x (1991). Article CAS PubMed Google Scholar * Dibattista, M. & Reisert, J. The odorant
receptor-dependent role of olfactory marker protein in olfactory receptor neurons. _J. Neurosci._ 36, 2995–3006. https://doi.org/10.1523/JNEUROSCI.4209-15.2016 (2016). Article CAS PubMed
PubMed Central Google Scholar * Reisert, J. & Restrepo, D. Molecular tuning of odorant receptors and its implication for odor signal processing. _Chem. Senses_ 34, 535–545.
https://doi.org/10.1093/chemse/bjp028 (2009). Article CAS PubMed PubMed Central Google Scholar * Saito, H., Chi, Q., Zhuang, H., Matsunami, H. & Mainland, J. D. Odor coding by a
mammalian receptor repertoire. _Sci. Signal_ 2, ra9. https://doi.org/10.1126/scisignal.2000016 (2009). Article PubMed PubMed Central Google Scholar * Mainland, J. D., Li, Y. R., Zhou,
T., Liu, W. L. & Matsunami, H. Human olfactory receptor responses to odorants. _Sci. Data_ 2, 150002. https://doi.org/10.1038/sdata.2015.2 (2015). Article CAS PubMed PubMed Central
Google Scholar * Pelosi, P. Odorant-binding proteins: Structural aspects. _Ann. N. Y. Acad. Sci._ 855, 281–293 (1998). Article ADS CAS PubMed Google Scholar * Debat, H. _et al._
Identification of human olfactory cleft mucus proteins using proteomic analysis. _J. Proteome Res._ 6, 1985–1996. https://doi.org/10.1021/pr0606575 (2007). Article CAS PubMed Google
Scholar * Heydel, J. M. _et al._ Odorant-binding proteins and xenobiotic metabolizing enzymes: Implications in olfactory perireceptor events. _Anat. Rec. (Hoboken)_ 296, 1333–1345.
https://doi.org/10.1002/ar.22735 (2013). Article CAS PubMed Google Scholar * Ijichi, C. _et al._ Metabolism of odorant molecules in human nasal/oral cavity affects the odorant
perception. _Chem. Senses_ 44, 465–481. https://doi.org/10.1093/chemse/bjz041 (2019). Article CAS PubMed Google Scholar * Heydel, J. M., Faure, P. & Neiers, F. Nasal odorant
metabolism: Enzymes, activity and function in olfaction. _Drug Metab. Rev._ 51, 224–245. https://doi.org/10.1080/03602532.2019.1632890 (2019). Article CAS PubMed Google Scholar *
Boichot, V. _et al._ The role of perireceptor events in flavor perception. _Front. Food Sci. Technol._ https://doi.org/10.3389/frfst.2022.989291 (2022). Article Google Scholar *
Kornbausch, N., Debong, M. W., Buettner, A., Heydel, J. M. & Loos, H. M. Odorant metabolism in humans. _Angew. Chem. Int. Ed. Engl._ 61, e202202866.
https://doi.org/10.1002/anie.202202866 (2022). Article CAS PubMed PubMed Central Google Scholar * Robert-Hazotte, A. _et al._ Nasal odorant competitive metabolism is involved in the
human olfactory process. _J. Agric. Food Chem._ 70, 8385–8394. https://doi.org/10.1021/acs.jafc.2c02720 (2022). Article CAS PubMed Google Scholar * Schwartz, M. _et al._ Oral enzymatic
detoxification system: Insights obtained from proteome analysis to understand its potential impact on aroma metabolization. _Compr. Rev. Food Sci. Food Saf._ 20, 5516–5547.
https://doi.org/10.1111/1541-4337.12857 (2021). Article CAS PubMed Google Scholar * Kuhlmann, K. _et al._ The membrane proteome of sensory cilia to the depth of olfactory receptors.
_Mol. Cell Proteom._ 13, 1828–1843. https://doi.org/10.1074/mcp.M113.035378 (2014). Article CAS Google Scholar * Heydel, J. M. _et al._ Characterization of rat glutathione transferases in
olfactory epithelium and mucus. _PLoS ONE_ https://doi.org/10.1371/journal.pone.0220259 (2019). Article PubMed PubMed Central Google Scholar * Schwartz, M. _et al._ Interactions between
odorants and glutathione transferases in the human olfactory cleft. _Chem. Senses_ 45, 645–654. https://doi.org/10.1093/chemse/bjaa055 (2020). Article CAS PubMed Google Scholar *
Lazard, D. _et al._ Odorant signal termination by olfactory UDP glucuronosyl transferase. _Nature_ 349, 790–793. https://doi.org/10.1038/349790a0 (1991). Article ADS CAS PubMed Google
Scholar * Nagashima, A. & Touhara, K. Enzymatic conversion of odorants in nasal mucus affects olfactory glomerular activation patterns and odor perception. _J. Neurosci._ 30,
16391–16398. https://doi.org/10.1523/JNEUROSCI.2527-10.2010 (2010). Article CAS PubMed PubMed Central Google Scholar * Munoz-Gonzalez, C., Feron, G., Brule, M. & Canon, F.
Understanding the release and metabolism of aroma compounds using micro-volume saliva samples by ex vivo approaches. _Food Chem._ 240, 275–285. https://doi.org/10.1016/j.foodchem.2017.07.060
(2018). Article CAS PubMed Google Scholar * Ijichi, C. _et al._ Odorant metabolism of the olfactory cleft mucus in idiopathic olfactory impairment patients and healthy volunteers. _Int.
Forum Allergy Rhinol._ https://doi.org/10.1002/alr.22897 (2021). Article PubMed Google Scholar * Muñoz-González, C., Brule, M., Martin, C., Feron, G. & Canon, F. Molecular mechanisms
of aroma persistence: From noncovalent interactions between aroma compounds and the oral mucosa to metabolization of aroma compounds by saliva and oral cells. _Food Chem._ 373, 131467.
https://doi.org/10.1016/j.foodchem.2021.131467 (2022). Article CAS PubMed Google Scholar * Muñoz-González, C., Feron, G., Brulé, M. & Canon, F. Understanding the release and
metabolism of aroma compounds using micro-volume saliva samples by ex vivo approaches. _Food Chem._ 240, 275–285. https://doi.org/10.1016/j.foodchem.2017.07.060 (2018). Article CAS PubMed
Google Scholar * Ployon, S., Brulé, M., Andriot, I., Morzel, M. & Canon, F. Understanding retention and metabolization of aroma compounds using an in vitro model of oral mucosa. _Food
Chem._ 318, 126468. https://doi.org/10.1016/j.foodchem.2020.126468 (2020). Article CAS PubMed Google Scholar * Ployon, S., Brule, M., Andriot, I., Morzel, M. & Canon, F.
Understanding retention and metabolization of aroma compounds using an in vitro model of oral mucosa. _Food Chem._ 318, 126468. https://doi.org/10.1016/j.foodchem.2020.126468 (2020). Article
CAS PubMed Google Scholar * Takaoka, N. _et al._ Involvement of aldehyde oxidase in the metabolism of aromatic and aliphatic aldehyde-odorants in the mouse olfactory epithelium. _Arch.
Biochem. Biophys._ 715, 109099. https://doi.org/10.1016/j.abb.2021.109099 (2022). Article CAS PubMed Google Scholar * Yoshikawa, K. _et al._ The human olfactory cleft mucus proteome and
its age-related changes. _Sci. Rep._ 8, 17170. https://doi.org/10.1038/s41598-018-35102-2 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * O’Connor, T., Ireland, L. S.,
Harrison, D. J. & Hayes, J. D. Major differences exist in the function and tissue-specific expression of human aflatoxin B1 aldehyde reductase and the principal human aldo-keto reductase
AKR1 family members. _Biochem. J._ 343(Pt 2), 487–504 (1999). Article CAS PubMed PubMed Central Google Scholar * Palackal, N. T., Burczynski, M. E., Harvey, R. G. & Penning, T. M.
Metabolic activation of polycyclic aromatic hydrocarbon trans-dihydrodiols by ubiquitously expressed aldehyde reductase (AKR1A1). _Chem. Biol. Interact._ 130–132, 815–824.
https://doi.org/10.1016/s0009-2797(00)00237-4 (2001). Article PubMed Google Scholar * Liang, L. P. & Patel, M. Mitochondrial oxidative stress and increased seizure susceptibility in
Sod2(–/+) mice. _Free Radic. Biol. Med._ 36, 542–554. https://doi.org/10.1016/j.freeradbiomed.2003.11.029 (2004). Article CAS PubMed Google Scholar * Seeley, S. K. _et al._ Metabolism of
oxidized linoleic acid by glutathione transferases: Peroxidase activity toward 13-hydroperoxyoctadecadienoic acid. _Biochim. Biophys. Acta_ 1760, 1064–1070.
https://doi.org/10.1016/j.bbagen.2006.02.020 (2006). Article CAS PubMed Google Scholar * Heydel, J. M. _et al._ Characterization of rat glutathione transferases in olfactory epithelium
and mucus. _PLoS ONE_ 14, e0220259. https://doi.org/10.1371/journal.pone.0220259 (2019). Article CAS PubMed PubMed Central Google Scholar * Vorachek, W. R., Pearson, W. R. & Rule,
G. S. Cloning, expression, and characterization of a class-mu glutathione transferase from human muscle, the product of the GST4 locus. _Proc. Natl. Acad. Sci. USA_ 88, 4443–4447.
https://doi.org/10.1073/pnas.88.10.4443 (1991). Article ADS CAS PubMed PubMed Central Google Scholar * Robert-Hazotte, A. _et al._ Nasal mucus glutathione transferase activity and
impact on olfactory perception and neonatal behavior. _Sci. Rep._ 9, 3104. https://doi.org/10.1038/s41598-019-39495-6 (2019). Article ADS CAS PubMed PubMed Central Google Scholar *
Schwartz, M. _et al._ Role of human salivary enzymes in bitter taste perception. _Food Chem._ 386, 132798. https://doi.org/10.1016/j.foodchem.2022.132798 (2022). Article CAS PubMed Google
Scholar * Jensen-Jarolim, E., Pacios, L. F., Bianchini, R., Hofstetter, G. & Roth-Walter, F. Structural similarities of human and mammalian lipocalins, and their function in innate
immunity and allergy. _Allergy_ 71, 286–294. https://doi.org/10.1111/all.12797 (2016). Article CAS PubMed Google Scholar * Tegoni, M. _et al._ Mammalian odorant binding proteins.
_Biochim. Biophys. Acta_ 1482, 229–240. https://doi.org/10.1016/s0167-4838(00)00167-9 (2000). Article CAS PubMed Google Scholar * Martin, H. J. & Maser, E. Role of human
aldo-keto-reductase AKR1B10 in the protection against toxic aldehydes. _Chem. Biol. Interact._ 178, 145–150. https://doi.org/10.1016/j.cbi.2008.10.021 (2009). Article CAS PubMed Google
Scholar * Nakashima, T., Kimmelman, C. P. & Snow, J. B. Jr. Structure of human fetal and adult olfactory neuroepithelium. _Arch. Otolaryngol._ 110, 641–646.
https://doi.org/10.1001/archotol.1984.00800360013003 (1984). Article CAS PubMed Google Scholar * Morrison, E. E. & Costanzo, R. M. Morphology of the human olfactory epithelium. _J.
Comp. Neurol._ 297, 1–13. https://doi.org/10.1002/cne.902970102 (1990). Article CAS PubMed Google Scholar * Moran, D. T., Rowley, J. C. 3rd., Jafek, B. W. & Lovell, M. A. The fine
structure of the olfactory mucosa in man. _J. Neurocytol._ 11, 721–746. https://doi.org/10.1007/BF01153516 (1982). Article CAS PubMed Google Scholar * Wierzchowski, J., Pietrzak, M.,
Szelag, M. & Wroczynski, P. Salivary aldehyde dehydrogenase-reversible oxidation of the enzyme and its inhibition by caffeine, investigated using fluorimetric method. _Arch. Oral Biol._
53, 423–428. https://doi.org/10.1016/j.archoralbio.2007.11.004 (2008). Article CAS PubMed Google Scholar * Hsu, L. C., Chang, W. C., Hiraoka, L. & Hsieh, C. L. Molecular cloning,
genomic organization, and chromosomal localization of an additional human aldehyde dehydrogenase gene, ALDH6. _Genomics_ 24, 333–341. https://doi.org/10.1006/geno.1994.1624 (1994). Article
CAS PubMed Google Scholar * Luo, D. _et al._ Heat shock protein 90-alpha mediates aldo-keto reductase 1B10 (AKR1B10) protein secretion through secretory lysosomes. _J. Biol. Chem._ 288,
36733–36740. https://doi.org/10.1074/jbc.M113.514877 (2013). Article CAS PubMed PubMed Central Google Scholar * Solobodowska, S., Giebultowicz, J., Wolinowska, R. & Wroczynski, P.
Contribution of ALDH1A1 isozyme to detoxification of aldehydes present in food products. _Acta Pol. Pharm._ 69, 1380–1383 (2012). CAS PubMed Google Scholar * Pappa, A., Chen, C.,
Koutalos, Y., Townsend, A. J. & Vasiliou, V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. _Free Radic. Biol. Med._
34, 1178–1189. https://doi.org/10.1016/s0891-5849(03)00070-4 (2003). Article CAS PubMed Google Scholar * Khanna, M. _et al._ Discovery of a novel class of covalent inhibitor for aldehyde
dehydrogenases. _J. Biol. Chem._ 286, 43486–43494. https://doi.org/10.1074/jbc.M111.293597 (2011). Article CAS PubMed PubMed Central Google Scholar * Surburg, H. & Panten, J.
_Common Fragrance and Flavor Materials: Preparation, Properties and Uses. _5th Ed. (completely revised and enlarged edition) (2006). * Feron, V. J. _et al._ Aldehydes: occurrence,
carcinogenic potential, mechanism of action and risk assessment. _Mutat. Res._ 259, 363–385. https://doi.org/10.1016/0165-1218(91)90128-9 (1991). Article ADS CAS PubMed Google Scholar *
Giebultowicz, J., Wroczynski, P., Kosinski, P. & Pietrzak, B. The activity of salivary aldehyde dehydrogenase during the menstrual cycle and pregnancy. _Arch. Oral Biol._ 58, 261–265.
https://doi.org/10.1016/j.archoralbio.2012.11.005 (2013). Article CAS PubMed Google Scholar * Dalleau, S., Baradat, M., Gueraud, F. & Huc, L. Cell death and diseases related to
oxidative stress: 4-hydroxynonenal (HNE) in the balance. _Cell Death Differ._ 20, 1615–1630. https://doi.org/10.1038/cdd.2013.138 (2013). Article CAS PubMed PubMed Central Google Scholar
* LoPachin, R. M. & Gavin, T. Molecular mechanisms of aldehyde toxicity: A chemical perspective. _Chem. Res. Toxicol._ 27, 1081–1091. https://doi.org/10.1021/tx5001046 (2014). Article
CAS PubMed PubMed Central Google Scholar * Gaun, V., Martens, J. R. & Schwob, J. E. Lifespan of mature olfactory sensory neurons varies with location in the mouse olfactory
epithelium and age of the animal. _J. Comp. Neurol._ 530, 2238–2251. https://doi.org/10.1002/cne.25330 (2022). Article CAS PubMed Google Scholar * Shipley, M. T., Ennis, M. & Puche,
A. C. _The Rat Nervous System_. 3rd Ed. 923–964 (2004). * Tsuchida, S. & Yamada, T. _Glutathione Transferases_. https://doi.org/10.1016/B978-0-12-801238-3.04351-8 (Elsevier, 2014). *
Marchitti, S. A., Brocker, C., Stagos, D. & Vasiliou, V. Non-P450 aldehyde oxidizing enzymes: The aldehyde dehydrogenase superfamily. _Expert Opin. Drug Metab. Toxicol._ 4, 697–720.
https://doi.org/10.1517/17425255.4.6.697 (2008). Article CAS PubMed PubMed Central Google Scholar * Penning, T. M. & Drury, J. E. Human aldo-keto reductases: Function, gene
regulation, and single nucleotide polymorphisms. _Arch. Biochem. Biophys._ 464, 241–250. https://doi.org/10.1016/j.abb.2007.04.024 (2007). Article CAS PubMed PubMed Central Google
Scholar * Munoz-Gonzalez, C., Brule, M., Martin, C., Feron, G. & Canon, F. Molecular mechanisms of aroma persistence: From noncovalent interactions between aroma compounds and the oral
mucosa to metabolization of aroma compounds by saliva and oral cells. _Food Chem._ 373, 131467. https://doi.org/10.1016/j.foodchem.2021.131467 (2022). Article CAS PubMed Google Scholar *
Marchal, S. & Branlant, G. Evidence for the chemical activation of essential cys-302 upon cofactor binding to nonphosphorylating glyceraldehyde 3-phosphate dehydrogenase from
_Streptococcus mutans_. _Biochemistry_ 38, 12950–12958. https://doi.org/10.1021/bi990453k (1999). Article CAS PubMed Google Scholar * Sreerama, L., Hedge, M. W. & Sladek, N. E.
Identification of a class 3 aldehyde dehydrogenase in human saliva and increased levels of this enzyme, glutathione _S_-transferases, and DT-diaphorase in the saliva of subjects who
continually ingest large quantities of coffee or broccoli. _Clin. Cancer Res._ 1, 1153–1163 (1995). CAS PubMed Google Scholar * Giebultowicz, J. _et al._ Salivary aldehyde
dehydrogenase—Temporal and population variability, correlations with drinking and smoking habits and activity towards aldehydes contained in food. _Acta Biochim. Pol._ 57, 361–368 (2010).
Article CAS PubMed Google Scholar * Kurkivuori, J. _et al._ Acetaldehyde production from ethanol by oral streptococci. _Oral Oncol._ 43, 181–186.
https://doi.org/10.1016/j.oraloncology.2006.02.005 (2007). Article CAS PubMed Google Scholar * Schwartz, M., Canon, F., Feron, G., Neiers, F. & Gamero, A. Impact of oral microbiota
on flavor perception: From food processing to in-mouth metabolization. Foods https://doi.org/10.3390/foods10092006 (2021). * Neiers, F., Gourrat, K., Canon, F. & Schwartz, M. Metabolism
of cysteine conjugates and production of flavor sulfur compounds by a carbon-sulfur lyase from the oral anaerobe _Fusobacterium __nucleatum_. _J. Agric. Food Chem._ 70, 9969–9979.
https://doi.org/10.1021/acs.jafc.2c01727 (2022). Article CAS PubMed Google Scholar * Kawafune, K. _et al._ Strong association between the 12q24 locus and sweet taste preference in the
Japanese population revealed by genome-wide meta-analysis. _J. Hum. Genet._ 65, 939–947. https://doi.org/10.1038/s10038-020-0787-x (2020). Article CAS PubMed Google Scholar * Bouyssie,
D. _et al._ Proline: An efficient and user-friendly software suite for large-scale proteomics. _Bioinformatics_ 36, 3148–3155. https://doi.org/10.1093/bioinformatics/btaa118 (2020). Article
CAS PubMed PubMed Central Google Scholar * Kabsch, W. Xds. _Acta Crystallogr. D Biol. Crystallogr._ 66, 125–132. https://doi.org/10.1107/S0907444909047337 (2010). Article CAS PubMed
PubMed Central Google Scholar * Winn, M. D. _et al._ Overview of the CCP4 suite and current developments. _Acta Crystallogr. D Biol. Crystallogr._ 67, 235–242.
https://doi.org/10.1107/S0907444910045749 (2011). Article CAS PubMed PubMed Central Google Scholar * Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. _Acta
Crystallogr. D Biol. Crystallogr._ 60, 2126–2132. https://doi.org/10.1107/S0907444904019158 (2004). Article CAS PubMed Google Scholar * Adams, P. D. _et al._ PHENIX: A comprehensive
Python-based system for macromolecular structure solution. _Acta Crystallogr. D Biol. Crystallogr._ 66, 213–221. https://doi.org/10.1107/S0907444909052925 (2010). Article CAS PubMed
PubMed Central Google Scholar * Chen, V. B. _et al._ MolProbity: All-atom structure validation for macromolecular crystallography. _Acta Crystallogr. D Biol. Crystallogr._ 66, 12–21.
https://doi.org/10.1107/S0907444909042073 (2010). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS The authors want to acknowledge beamtime and staff assistance for
data collection at the synchrotron beamline PROXIMA1 (SOLEIL synchrotron, Saint-Aubin, France). FUNDING This study was funded by Agence Nationale de la Recherche (Grant Nos.
ANR-18-CE92-0018-01, ANR-20-CE21-0002, ANR-22-CE21-0001 and ANR-16-CE21-0004-01). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Flavour Perception: Molecular Mechanisms (Flavours), INRAE,
CNRS, Institut Agro, Université de Bourgogne Franche-Comté, Dijon, France Valentin Boichot, Franck Menetrier, Francis Canon, Jean-Marie Heydel, Mathieu Schwartz & Fabrice Neiers * CNRS,
Inserm, CHU Lille, Institut Pasteur de Lille, UAR CNRS 2014-US Inserm 41-PLBS, University of Lille, Lille, France Jean-Michel Saliou * UMR 1231, Lipides Nutrition Cancer, INSERM, 21000,
Dijon, France Frederic Lirussi * UFR des Sciences de Santé, Université Bourgogne Franche-Comté, 25000, Besançon, France Frederic Lirussi * Plateforme PACE, Laboratoire de
Pharmacologie-Toxicologie, Centre Hospitalo-Universitaire Besançon, 25000, Besançon, France Frederic Lirussi * Department of Otolaryngology-Head and Neck Surgery, Dijon University Hospital,
21000, Dijon, France Mireille Folia * Smell and Taste Clinic, Department of Otorhinolaryngology, TU Dresden, Dresden, Germany Thomas Hummel & Susanne Menzel * Chair of Tissue Engineering
and Regenerative Medicine, University Hospital Wuerzburg, Roentgenring 11, 97070, Wuerzburg, Germany Maria Steinke * Fraunhofer Institute for Silicate Research ISC, Roentgenring 11, 97070,
Wuerzburg, Germany Maria Steinke * Department of Otorhinolaryngology-Head and Neck Surgery, RWTH Aachen University Hospital, Aachen, Germany Stephan Hackenberg Authors * Valentin Boichot
View author publications You can also search for this author inPubMed Google Scholar * Franck Menetrier View author publications You can also search for this author inPubMed Google Scholar *
Jean-Michel Saliou View author publications You can also search for this author inPubMed Google Scholar * Frederic Lirussi View author publications You can also search for this author
inPubMed Google Scholar * Francis Canon View author publications You can also search for this author inPubMed Google Scholar * Mireille Folia View author publications You can also search for
this author inPubMed Google Scholar * Jean-Marie Heydel View author publications You can also search for this author inPubMed Google Scholar * Thomas Hummel View author publications You can
also search for this author inPubMed Google Scholar * Susanne Menzel View author publications You can also search for this author inPubMed Google Scholar * Maria Steinke View author
publications You can also search for this author inPubMed Google Scholar * Stephan Hackenberg View author publications You can also search for this author inPubMed Google Scholar * Mathieu
Schwartz View author publications You can also search for this author inPubMed Google Scholar * Fabrice Neiers View author publications You can also search for this author inPubMed Google
Scholar CONTRIBUTIONS V.B. performed the experiments and analysis and wrote the manuscript. F.M. performed the immunohistochemistry experiments. J.-M.S. performed the mass spectrometry
experiments, M. F., T.H. S.M., M. St, S.H., collected the human samples. F.C., F.L., J.-M.H. reviewed & edited the manuscript. F.N. and M.Sc. had a role in the conceptualization, funding
acquisition, investigation, methodology, project administration, and wrote the manuscript. CORRESPONDING AUTHORS Correspondence to Mathieu Schwartz or Fabrice Neiers. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published
maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution
4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and
the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's
Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not
permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Boichot, V., Menetrier, F., Saliou, JM. _et al._ Characterization of human
oxidoreductases involved in aldehyde odorant metabolism. _Sci Rep_ 13, 4876 (2023). https://doi.org/10.1038/s41598-023-31769-4 Download citation * Received: 15 December 2022 * Accepted: 16
March 2023 * Published: 25 March 2023 * DOI: https://doi.org/10.1038/s41598-023-31769-4 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
