Nuclear overexpression of dna damage-inducible transcript 4 (ddit4) is associated with aggressive tumor behavior in patients with pancreatic tumors
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT DNA damage-inducible transcript 4 (DDIT4) is induced in various cellular stress conditions. Several studies showed that the dysregulation of DDIT4 is involved in different
malignancies with paradoxical expressions and roles. Therefore, this study investigated the clinical significance, prognostic, and diagnostic value of DDIT4 in different types of pancreatic
tumors (PT). The expression of DDIT4 and long non-coding RNA (TPTEP1) in mRNA level was examined in 27 fresh PT samples using Real-time quantitative PCR (RT-qPCR). Moreover, 200
formalin-fixed paraffin-embedded PT tissues, as well as 27 adjacent normal tissues, were collected to evaluate the clinical significance, prognostic, and diagnosis value of DDIT4 expression
by immunohistochemistry (IHC) on tissue microarrays (TMA) slides. The results of RT-qPCR showed that the expression of DDIT4 in tumor samples was higher than in normal samples which was
associated with high tumor grade (_P_ = 0.015) and lymphovascular invasion (_P_ = 0.048). Similar to this, IHC findings for nucleus, cytoplasm, and membrane localization showed higher
expression of DDIT4 protein in PT samples rather than in nearby normal tissues. A statistically significant association was detected between a high level of nuclear expression of DDIT4
protein, and lymphovascular invasion (_P_ = 0.025), as well as advanced TNM stage (_P_ = 0.034) pancreatic ductal adenocarcinoma (PDAC) and in pancreatic neuroendocrine tumor (PNET),
respectively. In contrast, a low level of membranous expression of DDIT4 protein showed a significant association with advanced histological grade (_P_ = 0.011), margin involvement (_P_ =
0.007), perineural invasion (_P_ = 0.023), as well as lymphovascular invasion (_P_ = 0.005) in PDAC. No significant association was found between survival outcomes and expression of DDIT4 in
both types. It was found that DDIT4 has rational accuracy and high sensitivity as a diagnostic marker. Our results revealed a paradoxical role of DDIT4 expression protein based on the site
of nuclear and membranous expression. The findings of this research indicated that there is a correlation between elevated nuclear expression of DDIT4 and the advancement and progression of
disease in patients with PT. Conversely, high membranous expression of DDIT4 was associated with less aggressive tumor behavior in patients with PDAC. However, further studies into the
prognostic value and biological function of DDIT4 are needed in future studies. SIMILAR CONTENT BEING VIEWED BY OTHERS HIGH EXPRESSION OF DNA DAMAGE-INDUCIBLE TRANSCRIPT 4 (DDIT4) IS
ASSOCIATED WITH ADVANCED PATHOLOGICAL FEATURES IN THE PATIENTS WITH COLORECTAL CANCER Article Open access 01 July 2021 IDENTIFICATION OF DNA DAMAGE RESPONSE-RELATED GENES AS BIOMARKERS FOR
CASTRATION-RESISTANT PROSTATE CANCER Article Open access 10 November 2023 ALKBH5-MEDIATED M6A DEMETHYLATION OF KCNK15-AS1 INHIBITS PANCREATIC CANCER PROGRESSION VIA REGULATING KCNK15 AND
PTEN/AKT SIGNALING Article Open access 01 December 2021 INTRODUCTION Pancreatic tumor (PT), a highly aggressive malignancy, is predicted to become the second leading cause of cancer-related
deaths in the next decade1. Based on global cancer statistics 2020, the number of new cases and deaths of PT were 496,773 and 466,003, respectively, indicating an extremely fatal rate2.
There were several histological types of PT comprising pancreatic ductal adenocarcinoma (PDAC), the most frequent, and pancreatic neuroendocrine tumor (PNET), which arise from pluripotent
cells3,4. It has been found that the prevalence and incidence of PNET have gradually increased over the past 40 years and can be as high as 10% in autopsy studies5,6. The 5-year survival
rate of PT is roughly 10 percent across the world, with minimal change throughout the past few decades7,8. Due to the early detection restrictions, low respectability rate at the time of
diagnosis, early metastasis nature, and high resistance rate to neoadjuvant therapy, the prognosis is dismal9,10. Consequently, the need for prompt diagnosis and treatment of this insidious
cancer is recognized globally. The discovery of tumor markers is a valuable and promising tool that has the potential in order to diagnose patients at an early stage of the disease, improve
prognosis, and enhance treatment response11,12,13. DNA damage-inducible transcript 4 (DDIT4), also known as a regulated in development and DNA damage response 1 (REDD1) protein or
hypoxia-inducible factor 1 (HIF1)-responsive protein RTP801 (RTP801), was cloned in 200214,15. It is strongly upregulated under various cellular stresses, such as hypoxia16, heat shock17,
ionizing radiation18, and chemical molecules19. It has been indicated that DDIT4 inhibits the mammalian target of rapamycin complex 1 (mTORC1) and has a potential role in regulating cell
growth, tumorigenesis, and autophagy pathway20. A growing body of evidence has presented that dysregulation of DDIT4 occurs in diverse human cancers with contradictory roles as an oncogene
or tumor suppressor21,22,23. As an oncogene, DDIT4 can cause the occurrence, and development of cancer via a link with RAS, stabilizing HIF1, and reducing apoptotic rate through increased
anti-apoptotic proteins, which leads to an increase in cancer cell survival, proliferation, and migration and decreased apoptosis22,24,25,26,27,28,29,30. In accordance with this concept, an
increase in the expression of DDIT4 at both the mRNA and protein levels has been observed in several types of cancer, such as colorectal carcinoma (CRC)31,32, bladder urothelial carcinoma
(BUC)22, oral squamous cell carcinoma (OSCC)33, ovarian cancer (OC)28,34, and gastric cancer (GC)27. In contrast, several studies have demonstrated that DDIT4 is activated under diverse
stress conditions in cells, resulting in cell death through inhibiting mTORC116,35,36,37,38. Consequently, it appears that DDIT4 may act as a tumor-suppressing role in the progression of
cancer23,39,40,41. Thus, the role of DDIT4 in cancer promotion or suppression is still unclear42. Besides, in silico analysis indicated DDIT4 from the 3'-untranslated regions (UTR)
binds to TPTE pseudogene 1 (TPTEP1), a long non-coding RNA (lncRNA) gene, regulates gene expression31. LncRNAs are non-coding RNAs with more than 200 nucleotides in length and have critical
roles in existing cancers via gene transcription regulating, post-transcriptional protein activity control, or organizing nuclear domains43,44,45. Hence, in the present study, the mRNA
expression of DDIT4 and TPTEP1 in fresh PT samples and their adjacent normal tissues was assessed using RT-qPCR. Besides, for the first time, the expression levels and localization of DDIT4
protein were examined in the nucleus, cytoplasm, and membranous of PT tissues using the immunohistochemistry (IHC) method on tissue microarrays (TMAs) slides. Subsequently, the associations
between expression levels of DDIT4 at different subcellular locations and clinicopathological features, also survival outcomes were analyzed. RESULTS BIOINFORMATICS APPROACHES Based on the
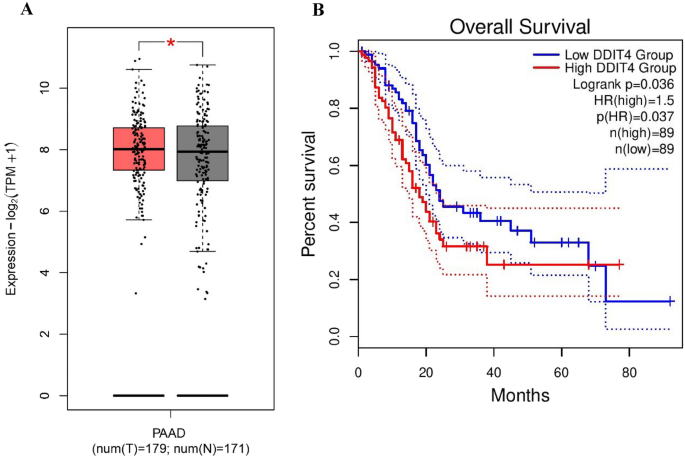
analysis conducted on the TCGA database through GEPIA2, few changes were observed in the mRNA levels of DDIT4 in pancreatic adenocarcinoma (PAAD) tissue samples compared to the normal
tissues (_P_ < 0.05, Fig. 1A). Furthermore, the prognostic value of DDIT4 in PAAD was assessed by the use of transcriptome sequencing data from the GEPIA2 database. As a result, we found
a high expression level of DDIT4 associated with the worse overall survival (OS) (HR (high) = 1.5, _P_ (HR) = 0.037) in PAAD patients (Fig. 1B). PATIENTS’ CHARACTERISTICS OF FRESH TISSUE
SAMPLES A total of 27 fresh tissue PDAC samples and 27 adjacent normal specimens were collected to investigate DDIT4 mRNA expression, of which 16 (59.2%) cases were males and 11 (40.8%) were
females (male/female ratio = 1.45). The median age of patients was 62 ± 10.18 years old, ranging from 29 to 79, and the median tumor size was 2.7 cm (range: 1–12 cm). Histological grade of
samples was available for 22 sample tissues, which are categorized into three groups as follows: 9 (40.9%) well-differentiated, 12 (54.5%) moderately differentiated, and 1 (4.6%) poorly
differentiated. Further, samples were divided into three stages (I-III): 13 (56.5%) stage I, 8 (34.7%) stage II, and 2 (8.8%) stage III. Margin involvement, lymphovascular, and perineural
invasion were observed in 3 (11.1%), 9 (33.3%), and 9 (33.3%), respectively. Lymph node metastasis was also present in 12 (44.4%) patients. EVALUATION OF DDIT4 AND TPTEP1 MRNA EXPRESSION
LEVEL IN FRESH TUMOR TISSUES OF PANCREATIC TUMOR AND THEIR ADJACENT NORMAL TISSUES The analysis of RT-qPCR data using the Mann–Whitney U test indicated that the mRNA expression levels of
DDIT4 were significantly increased in PT patients compared to the healthy group (_P_ < 0.05) (Fig. 2A). However, there was no significant difference between TPTEP1 mRNA expression in
tumors and normal samples (Fig. 2B). ASSOCIATIONS BETWEEN DDIT4 MRNA EXPRESSION AND CLINICOPATHOLOGICAL PARAMETERS IN FRESH TISSUE SAMPLES Kruskal–Walli’s and Mann–Whitney U test analyses
were performed to find any association between the median expression of the DDIT4 gene and the clinicopathological parameters of the PT patients. The findings of our study indicated a
statistically significant increase in the median expression levels of DDIT4 in patients with moderate/poor tumor differentiation in comparison to those with well-differentiated tumors (_P_ =
0.015) (Fig. 2C). Furthermore, as shown in frame D of Fig. 2, the median expression of DDIT4 was significantly higher in the patients with lymphovascular invasion than in patients without
lymphovascular invasion (_P_ = 0.048). There was no significant relationship between the median expression of DDIT4 and stage (I vs. II/III), as well as perineural invasion and lymph node
metastasis (Fig. 2E–G). CHARACTERISTICS OF PATIENTS’ FORMALIN-FIXED PARAFFIN-EMBEDDED (FFPE) TISSUE SAMPLES To examine the clinical significance and subcellular localization of DDIT4 at the
protein level, 200 FFPE tissue specimens were enrolled in this cross-sectional study, of whom 177 samples were PDAC, and the others were PNET type. The median age of total samples was 60
years old, ranging from 12 to 85. Among all patients, 108 (54.0%) cases were male, and 92 (46.0%) were female. The histological grade of tumor cells was divided into three categories as
well, moderately, and weakly differentiated. All clinicopathological features of our total samples and types of PT are described in Table 1. EXPRESSION OF DDIT4 IN PANCREATIC TUMOR COMPARED
WITH ADJACENT NORMAL SAMPLES The protein expression levels of DDIT4 were evaluated using the IHC technique on TMA sections by three distinct scoring methods that comprise the intensity of
staining, percentage of positive tumor cells, and H-score. DDIT4 was expressed at different intensities in the nucleus, cytoplasm, and membranous in two types of PT as well as adjacent
normal samples (Fig. 3). The mean expression level (based on H-score) of DDIT4 was 80 in tumor samples (in all expression); however, the mean expression of DDIT4 was 20 in the nucleus, 30 in
the cytoplasm, and 32 in the membranous in adjacent normal tissue, indicating that the expression of DDIT4 in PTs was high compared to adjacent normal tissues (Table 2 and Fig. 4). Besides,
Mann–Whitney U tests showed a significant difference between the median nuclear, cytoplasmic, and membranous expression of DDIT4, and the median expression of adjacent normal tissues (_P_ =
0.012, _P_ = 0.001, and _P_ = 0.026, respectively) (Table 2). ASSOCIATIONS BETWEEN DDIT4 EXPRESSION (NUCLEUS, CYTOPLASM, AND MEMBRANE) AND CLINICOPATHOLOGICAL CHARACTERISTICS OF PDAC TISSUE
SAMPLES The association between the expression of DDIT4 protein and the clinicopathological parameters of PT was investigated using Pearson's χ2 test. Our analysis demonstrated a
significant association between the nuclear expression of DDIT4 protein and lymphovascular invasion (_P_ = 0.025). We also found a statistically significant association between a membranous
expression of DDIT4 protein and histological grade (_P_ = 0.011), margin involvement (_P_ = 0.007), perineural invasion (_P_ = 0.023), as well as a lymphovascular invasion (_P_ = 0.005).
There was no significant association between cytoplasmic expression of DDIT4 and clinicopathological features (Table 3). Besides, the correlation between DDIT4 protein expression and
clinicopathological characteristics was examined using spearman’s correlation tests. Our results indicated a significant direct correlation between a high level of nuclear expression of
DDIT4 and lymphovascular invasion (_P_ = 0.009). Furthermore, we observed an inverse correlation between increased membranous expression of DDIT4 protein and advanced histological grade (_P_
= 0.011), margin involvement (_P_ = 0.001), perineural invasion (_P_ = 0.007), as well as lymphovascular invasion (_P_ = 0.001). ASSOCIATIONS BETWEEN DDIT4 EXPRESSION (NUCLEUS, CYTOPLASM,
AND MEMBRANE) AND CLINICOPATHOLOGICAL CHARACTERISTICS OF PNET TISSUE SAMPLES Pearson's χ2 test exhibited a significant association between the nuclear expression of DDIT4 protein and
histological grade (_P_ = 0.034). Moreover, our Spearman’s correlation tests showed a direct correlation between the nuclear expression of DDIT4, and the high grade (_P_ = 0.014). We did not
find any association or correlation between cytoplasmic and membranous DDIT4 protein expressions and the clinicopathological parameters (Table 4). INFORMATION ON CLINICAL OUTCOMES IN TYPES
OF PT In the current study, from 200 patients, follow-up data for 10 patients were not available. Of them, 7 patients had PDAC, and 3 patients had PNET. PANCREATIC DUCTAL ADENOCARCINOMA
CANCER Among 190 patients, 33 (19.4%) showed tumor recurrence, 68 (40.0%) had metastasis, and 8 (4.7%) showed recurrence and metastasis. Cancer-related death occurred in 106 (62.3%)
patients. The mean OS/disease-specific survival (DSS) and progression-free survival (PFS) follow-up time for patients with high nuclear expression was shorter than the low nuclear expression
of DDIT4 (OS/DSS = 17 and 20 months, respectively, and PFS = 13 and 16 months, respectively). However, the mean OS/DSS and PFS follow-up time for patients with high cytoplasmic, and
membranous expression were longer than the low expression of DDIT4 (cytoplasmic: OS/DSS = 20 and 17 months, and PFS = 16 and 13 months, respectively; membranous: OS/DSS = 19 and 18 months,
and PFS = 15 and 14 months, respectively); indicating that high nuclear expression has shorter survival than low expression; while high cytoplasmic and membranous expression of DDIT4 was
related to longer survival. The main characteristics of survival outcomes are exhibited in Table 5. PANCREATIC NEUROENDOCRINE TUMOR In this type of PT, tumor recurrence and metastasis
occurred in 2 (10.0) and 4 (20.0) patients, respectively. Throughout follow-up, all death cases died of pancreatic tumor. The median and mean OS and DSS time were 27 months (Q1 = 18 and Q3 =
37) and 27 (SD = 12.92), respectively. Additionally, the median and mean follow-up periods for PFS were 25 (Q1 = 17, Q3 = 35) and 24 (SD = 14.03) months, respectively. Patients with high
nuclear DDIT4 expression have been reported to have lower mean OS, DSS and PFS survival, which is consistent with PDAC survival data. However, there is not any difference between the mean
OS/DSS/PFS and high/low cytoplasmic and membranous expression. SURVIVAL ANALYSIS BASED ON THE EXPRESSION OF DDIT4 IN PT TYPES PANCREATIC DUCTAL ADENOCARCINOMA CANCER Kaplan–Meier survival
analysis exhibited no statistically significant differences between OS/DSS/PFS and the patients with high /low expression of DDIT4 (Fig. 5). Univariate and multivariate Cox regression
analyses were performed to evaluate the clinical significance of potential prognostic factors for OS, DSS, and PFS. On univariate analyses, tumor size, TNM stage, and distant metastasis were
detected as potential prognostic factors for OS, DSS, and PFS. In addition, age and lymphovascular invasion have a prognostic role for OS/PFS and DSS, respectively. Statistically
significant univariate analyses were incorporated into multivariate Cox regression analysis, in which the results indicated that distant metastasis was an independent risk factor affecting
OS (HR: 187; 95% CI 1.479–3.233; _P_ < 0.001), DSS (HR: 2.286; 95% CI 1.528–3.421; _P_ < 0.001), and PFS (HR: 2.680; 95% CI 1.819–3.949; _P < 0.001_, respectively) (Table 6).
PANCREATIC NEUROENDOCRINE TUMOR Kaplan–Meier survival analysis indicated that DDIT4 expression was not a prognostic factor in OS (Log-rank test; nuclear: _P_ = 0.812, cytoplasmic: _P_ =
0.955, membranous: _P_ = 0.603), DSS (Log-rank test; nuclear: _P_ = 0.812, cytoplasmic: _P_ = 0.955, membranous: _P_ = 0.603), and PFS (Log-rank test; nuclear: _P_ = 0.563, cytoplasmic: _P_
= 0.967, membranous: _P_ = 0.524). Besides, the results of univariate and multivariate Cox regression analyses showed that the clinicopathologic parameters were not significant factors
affecting OS, DSS, or PFS in the patients with PNET. DIAGNOSTIC VALUE OF THE DDIT4 IN PT VERSUS ADJACENT NORMAL TISSUES ROC curves, and the AUC of the expression level of DDIT4 protein
(nuclear, cytoplasmic, and membranous) were performed to explore the diagnostic value of the marker in discriminating pancreatic tumor from adjacent normal tissues (Fig. 6). The results of
ROC curves demonstrated an AUC of 0.64 (95% CI 0.53–0.75), with 68% sensitivity, and 57% specificity for nuclear expression of DDIT4 in PTs versus adjacent normal tissues. For cytoplasmic
expression of DDIT4, an AUC, sensitivity, as well as specificity were 0.70 (95% CI 0.59–0.80), 66%, and 75%, respectively. The membranous expression of DDIT4 also had an AUC, sensitivity,
and specificity of 0.62 (95% CI 0.53–0.71), 63%, and 50%, respectively. Table 7 displays the overall diagnostic results for DDIT4 expression. DISCUSSION Pancreatic tumor is recognized as the
most fatal and aggressive malignancy among other gastrointestinal cancers, which is estimated to become the second leading cause of cancer-related deaths by 20301,46. PT still has a high
death rate, with a 5-year survival rate of around 10%, despite significant advancements and notable improvements in the detection and treatment of many malignancies7. By the time of
diagnosis, PT often presents at an advanced stage of the disease, and has often metastasized to other organs47,48. As a result, it seems sensible to discover innovative biomarkers for early
detection, monitoring of recurrence and metastasis precisely, and the reduction of death related to the cancer. DDIT4 is a main coding RNA whose expressions are altered under the effect of
stressor factors, such as endoplasmic reticulum stress, hypoxia, heat shock, and ionizing radiation20,49. Research evidence showed that DDIT4 expression was dysregulated in various
cancers50,51. The multifaceted involvement of DDIT4 in angiogenesis, cancer medication therapy, and the regulation of intricate intercellular signaling networks has led to the proposition
that this molecule may possess a dualistic function in the initiation and advancement of cancer27,35,38,39,52. Notably, previous studies revealed that DDIT4 has cancer stem cell (CSC)
traits, which include self-renewal properties, quiescence, and dysregulated DNA repair31,38,53,54,55. CSCs, which are a subpopulation of cancer cells with self-renewal properties, lead to
the maintenance of tumors by generating differentiated tumor cells56,57,58,59. These features or characteristics of CSCs play crucial roles in heterogeneity, resistance to treatment,
recurrence, and metastasis of tumors56,57,58. In addition, the elevated levels of DDIT4 expression increase the survival and growth of tumor cells through stabilizing HIF1α, phosphorylation
of P53, and the inhibition of apoptosis pathways, and could also be related to the expression of stem cell markers27,29,30. There was merely one study addressing the clinical significance of
DDIT4 in pancreatic tumor, which showed that mutations in the 3'-UTR region of DDIT4 mRNA may affect autophagy by regulating the expression level of DDIT4 in PDAC tissues49. Therefore,
the expression and function of DDIT4 in PT still need to be clarified. Thus, this is the first study that evaluates the expression, prognostic, and diagnostic value of DDIT4 in both mRNA
and protein levels in either PDAC or PNET forms of pancreatic tumor. Consistent with the result of the bioinformatics analysis, which demonstrated increased mRNA expression of DDIT4, our
results in fresh tissue samples showed that mRNA levels of DDIT4 significantly increased in the pancreatic cancer patients when compared to adjacent normal tissues and had a positive
correlation with the high grade of the cancer and also with invasion of the lymphovascular. These findings align with the study conducted by Fattahi et al., which showed that the
upregulation of DDIT4 in colorectal cancer stem cell-enriched spheroids contributed to tumor advancement and metastasis31. Also, upregulation of DDIT4 was observed in GC tissues, which
promotes proliferation and tumorigenesis50. These data show that increased mRNA expression of DDIT4 is substantially related to more aggressive tumor behavior. In continuation, our research
results indicate a considerable upregulation of DDIT4 protein expression in malignant cells as compared to the adjacent normal samples. Our findings are in agreement with prior studies
showing the increased expression of DDIT4 protein in the nucleus, cytoplasm, and membranous tissues in various types of cancer22,28,31,32,41,50,60,61. It has also been reported that DDIT4 is
expressed mainly in the cell membrane and cytoplasm of normal cells, while it is more expressed in the nucleus of tumor cells20,62, suggesting that translocation of the molecule from the
membrane into the nucleus may be a risk factor for DDIT4 function changes that lead to oncogenes and may be associated with poor differentiation22. Our results confirmed this hypothesis
showed that the increased nuclear level of DDIT4 has a positive correlation with invasion to the lymphovascular tissues in PDAC and a high grade of the disease in the PNET samples. Both
results revealed that DDIT4 can be considered a risk factor for the progression of pancreatic tumor and plays an oncogenic role in the pancreas. In parallel with the findings, Wei et al.
reported that the DDIT4 gene has a p53 transcription-factor binding site, thus it may play a critical role in the p53-dependent tumorigenesis in the nucleus63. Similarly, other studies have
stated this fact that the high nuclear expression of DDIT4 protein is associated with advanced histological grade and enhances cancer proliferation and tumorigenesis32,33,64. To confirm the
results, a review article by Ding, F., and colleagues collected the recent information regarding the roles played by DDIT4 in the progression of tumors and proved that the nucleus expression
of DDIT4 is associated with tumor deterioration42. Therefore, DDIT4's functionality may be dependent on its precise subcellular location inside pancreatic cells. Notably, higher levels
of DDIT4 nuclear expression have been associated with more aggressive tumor behavior. Moreover, although there were no significant differences between the expression of DDIT4 and survival
in either PDAC or PNET patients, the pancreatic cancer patients with the high nuclear expression of DDIT4 had shorter survival duration compared to those with the low nuclear expression of
DDIT4, which was consistent with our clinicopathological findings. Our findings are parallel to the previous studies, concluding that DDIT4 expression is not a prognostic factor for
tumors32,50. Besides, our survival analysis demonstrated that the patients with high cytoplasmic and membranous expression of DDIT4 had a higher survival rate in PDAC cases. Besides, our
survival analysis showed that the patients with high cytoplasmic and membranous expression of DDIT4 had a higher survival rate in PDAC cases. However, there was no difference in DSS or PFS
follow-up months between PNET patients with high cytoplasmic or membranous expression of DDIT4 and low cytoplasmic or membranous expression. While cytoplasmic overexpression of DDIT4 protein
was shown in several studies to be a poor prognosis factor for tumor progression in ovarian carcinoma, acute myeloid leukemia, OSCC, and BUC22,28,33,65. Moreover, in this study, distant
metastasis was indicated as an independent prognostic factor for OS, DSS, and RFS in PDAC patients. Previous studies reported that metastasis has been significantly associated with disease
progression as well as unfavorable outcomes66,67. Distant metastasis has been considered as the major cause of the high death rate in patients with PT68. As well, in terms of diagnosis, it
has been shown that less than twenty percent of pancreatic cancer patients can be surgically removed at the time of their initial diagnosis69. Currently, carbohydrate antigen 19–9 (CA19-19),
which is extensively used in clinical practice, has been authorized by the US Food and Drug Administration (FDA) as the only biomarker for the detection of pancreatic tumor. Nevertheless,
the specificity of the current biomarker is limited, thus necessitating the urgent exploration and creation of new biomarkers70. Hence, we aimed to assess the diagnostic accuracy of DDIT4
and its sensitivity and specificity in discriminating PT from adjacent normal tissue samples. Our results revealed that the expression levels of DDIT4 protein proved that the molecule may be
considered the potential marker for diagnosis of pancreatic cancer patients with high sensitivity. Of note, these data support the direct role of DDIT4 in pancreatic tumors, which can be
used to diagnose the PT. However, we suggest assessing the concentration of DDIT4 in the serum of PT patients, a noninvasive detection method, as a potential diagnostic index, which might be
an innovative approach. Our study presents persuasive and consistent evidence regarding the clinical significance of DDIT4 in PT patients. However, we must recognize some limitations. These
include the relatively small number of PDAC and PNET samples utilized in the study, as well as the short duration of follow-up. These limitations may cast doubt on the prognostic role of
DDIT4. Therefore, it is crucial to evaluate the expression of DDIT4 in a larger sample size and extend the duration of follow-up to confirm the diagnostic and prognostic role of this marker.
Furthermore, we also acknowledge that we did not explore the detection mechanism and signaling pathways of DDIT4 in PT. To gain better insight into the mechanisms and functions of DDIT4 in
PT patients, it is advisable to determine the signaling pathways involved. CONCLUSION In summary, bioinformatics analysis confirmed that DDIT4 in mRNA level has increased in PT and can be
considered as a potential prognostic marker. Our findings showed that DDIT4 at the level of mRNA and protein was upregulated in the PT compared to the adjacent normal tissues. In contrast to
its cytoplasmic and membranous expression, higher nuclear level of DDIT4 is associated with more aggressive behavior, advanced disease, and a worse survival rate. Therefore, the nuclear
expression of DDIT4 could serve as a promising marker and a potential therapeutic target for PT patients. However, further investigations should be conducted regarding the molecular
mechanisms of DDIT4 in the progression of pancreatic tumor. MATERIALS AND METHODS DATA MINING THROUGH BIOINFORMATICS TOOLS (INVESTIGATION OF DDIT4 IN PATIENTS WITH PANCREATIC ADENOCARCINOMA)
Gene Expression Profiling Interactive Analysis (GEPIA2) is an online bioinformatics tool for expression data from RNA sequence for The Cancer Genome Atlas (TCGA) and the Genotype-Tissue
Expression (GTEx), tumor and normal samples71. In the primary search for the investigation of DDIT4 expression in sample tissues from patients with pancreatic adenocarcinoma, GEPIA2 was
performed. Moreover, GEPIA2 was applied to evaluate the prognostic potential with the overall survival curve of the DDIT4 expression in these samples which was stratified according to the
median expression of DDIT4 in GEPIA2. PATIENT’S CHARACTERISTICS AND SAMPLE COLLECTION In this cross-sectional study, 27 PDAC fresh tumor tissues and their adjacent normal tissue samples from
2020 to 2021 were collected from Firoozgar Hospital, a university-based hospital in Tehran, Iran. Patients who had undergone surgery without any prior chemotherapy/radiotherapy were
enrolled in this study. The samples were transferred to the laboratory, and frozen in liquid nitrogen. Patient information, including gender, age, TNM stage, histological grade,
lymphovascular invasion, perineural invasion, and lymph node metastasis were recorded. Moreover, 200 formalin-fixed paraffin-embedded (FFPE) tissue samples from PT patients were collected
from two university-based referral hospitals (Firoozgar and Imam Khomeini) in Tehran, Iran, between 2010 and 2021. These samples comprised two primary classifications of PT (PDAC and PNET)
that we encounter within our hospitals. None of the surgical participants received adjuvant therapy. Hematoxylin and Eosin (H & E) stained slides and the medical archival documents were
acquired to extract clinical and pathological parameters, comprising age, gender, tumor size (maximum tumor diameter), histological grade, TNM stage, distant metastasis, tumor recurrence,
margin involvement, macroscopic tumor extension, lymphovascular invasion, perineural invasion, and lymph node metastasis. However, it is important to note that chemotherapy was virtually
always given following surgery to all of our patients in accordance with recommendations, thus we did not include it as a variable in our analysis. Furthermore, 27 adjacent normal tissues
were utilized to determine the expression of DDIT4 compared to cancerous samples. OS represents the duration starting from the surgery date until the date of either death or the last
follow-up visit. DSS calculated from the date of surgery to the date of death related to the patient’s tumor and PFS defined as the date of the primary surgery and the last follow-up visit
for the patient with no evidence of disease, metastasis or recurrence, was also collected. Tumor staging was performed according to the pTNM classification for pancreatic tumor72. This study
received its ethical approval (Code: IR.IUMS.REC.1400.350.) from the Research Ethics Committee of Iran University of Medical Sciences. All procedures, including obtaining informed consent
from each human participant before surgery, were by the above-mentioned ethical standards. Thus, we confirm that all research was performed by relevant guidelines and regulations. RNA
EXTRACTION AND CDNA SYNTHESIS Total RNA was extracted from fresh tumor tissue samples, as well as normal adjacent tissues, using RNA Minipreps Super Kit (Bio Basic, Canada) according to the
manufacturer’s instructions. All extracted RNAs were measured in quantity and quality by Nanodrop (ThermoFisher Scientific, USA). Then RNA was reverse transcript into cDNA using the cDNA
Synthesis Kit (Yekta Tajhiz Azma, YT4500, Iran). REAL-TIME- QUANTITATIVE POLYMERASE CHAIN REACTION (RT‐QPCR) The specific primers (DDIT4, TPTEP1) for amplification with real-time-
quantitative PCR (RT-qPCR) were designed as described previously31. RT-qPCR was performed using the Rotor-Gene Q Light Cycler (Qiagen, Germany) by the SYBR Green qPCR master mix (Yekta
Tajhiz Azma, YT2551, Iran). After that, the mRNA level of the reference gene, GAPDH, was used as an internal control in each sample to normalize the relative expression levels of mRNAs.
RT-qPCR data were analyzed using the comparative Ct method (known as the 2-ΔΔCt method) to present relative expression levels of the genes73. The sequence-specific primers are shown in Table
8. TISSUE MICROARRAY (TMA) CONSTRUCTION Pancreatic tissue TMA blocks were constructed as described previously74,75. In brief, two experienced pathologists (B.B. and P.B) marked the three
most representative tumor areas in different parts of each block after matching them with corresponding H&E slides. The chosen tumor sections of each block were then punched out with a
0.6 mm diameter and placed into a fresh recipient paraffin block using a precision arraying tool (Tissue Arrayer Minicore; ALPHELYS, Plaisir, France). Next, completed TMA blocks were cut at
4-μm sections and transferred to adhesive slides. To avoid tumor heterogeneity as a significant concern during the TMA method and increase the accuracy and validity of the data analysis, TMA
blocks were constructed in three copies from each tumor specimen, and the mean scores of the three cores were calculated as the final score76,77,78. Notably, to compare the expression
patterns of DDIT4 with tumor tissue specimens, adjacent normal tissue samples were also included in each TMA block79. IMMUNOHISTOCHEMISTRY (IHC) STAINING DDIT4 expression was evaluated using
the IHC method as previously described80,81. First, all TMA sections were deparaffinized at 60 °C for 40 min and then rehydrated with xylene and graded ethylic alcohol. To block endogenous
peroxides and non-reactive staining, slides were treated with 3% H2O2 for 20 min at room temperature. After washing the slides in Tris Buffered Saline (TBS), antigens were retrieved by
immersing the tissue slides in citrate buffer (pH 6.0) for 10 min in an autoclave. Next, the slides were washed in TBS three times again and were stained by the primary antibody of
anti-DDIT4 (Biorbyt, Cambridge, UK) with a dilution of 1:80 overnight at 4 °C. Besides, rabbit immunoglobulin IgG (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) was utilized at the
same dilution of primary antibody for the isotype control test. The next day, following three times washing in TBS, TMA sections were treated with secondary antibody, including EnVision
TMMouse/Rabbit PolyVue HRP reagent (Dako; code K5007; Denmark), for 30 min at room temperature. Slides were incubated by 3,3′-diaminobenzidine (DAB) (Dako, Glostrup, Denmark) as a chromogen
and, after washing in TBS, counterstained by Mayer’s hematoxylin dye (Dako, Glostrup, Denmark) for 15 min to visualize the antigen. Eventually, the slides were dehydrated the same in graded
ethylic alcohol, cleared in xylene, and mounted for examination. Furthermore, TBS was utilized instead of the primary antibody as a negative control and normal liver tissue as a positive
control. EVALUATION OF IMMUNOSTAINING AND SCORING SCORE The staining of DDIT4 protein marker on TMA slides was scored by two independent pathologists (B.B and P.B.) using a semiquantitative
scoring system, blinded to the clinicopathological and survival parameters of the patients. To evaluate the expression of DDIT4, three scoring systems were used as follows: staining
intensity, the percentage of positive tumor cells, and the H-score. The intensity of staining was scored based on a 4-point scale (negative or non-staining = 0, weak = 1, moderate = 2, and
strong = 3), and the percentage of positive tumor cells was assessed and categorized into four groups (< 25, 25–50%, 51–75%, and > 75%). Then, the overall score was attained through
the Histochemical score (H-score) by multiplying the staining intensity and the percentage of the positive cells, which ranged from 0 to 300 in each case. The H-scores were classified into
two groups based on median: low expression (≤ median) and high expression (> median). STATISTICAL ANALYSIS All data were analyzed using "statistical software SPSS" version 25.0
(SPSS, Inc., IBM Corp, USA). The categorical and the quantitative data were represented by N (%) and mean (SD) or median (Q1, Q3), respectively. Pearson's chi-square and Spearman's
correlation tests were carried out to determine the significance of association and correlation between DDIT4 protein expression and clinicopathological characteristics. Kruskal–Wallis and
Mann–Whitney U tests were conducted to compare the study groups pairwise. Survival curves were plotted by applying the Kaplan–Meier method with a 95% confidence interval (CI) and compared
survival outcomes between low and high marker expression by the log-rank test. Furthermore, the univariate Cox proportional hazards regression model was utilized to determine which variables
impacted DSS or PFS. Then, those parameters that significantly affected the survival in univariate analysis were enrolled in multivariable Cox proportional hazards regression analyses. To
evaluate the diagnostic value of the DDIT4 protein, the area under the ROC curve (AUC), sensitivity, specificity, positive likelihood ratios (PLRs), and negative likelihood ratios (NLRs)
were calculated by using receiver operating characteristic (ROC) curve analysis. In all parts of the analysis, _P_ < _0.05_ was considered statistically significant difference. GraphPad
Prism version 8.4.3 software (GraphPad Software, La Jolla, CA, USA) was used for making the boxplots and scatterplots. ETHICAL APPROVAL All procedures performed in this study were in line
with the ethical standards of the institution at which this study was conducted. The Research Ethics Committee of Iran University of Medical Sciences issued IR.IUMS.REC.1400.350 for this
study. INFORMED CONSENT Informed consent was obtained from all individual participants, parents or legally authorized representatives of participants under legal age years old at the time of
sample collection with routine consent forms. DATA AVAILABILITY The analyzed data during the current study are available from the corresponding author on reasonable request. REFERENCES *
Rahib, L. _et al._ Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. _Cancer Res._ 74(11), 2913–2921 (2014).
Article CAS PubMed Google Scholar * Sung, H. _et al._ Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. _CA Cancer
J. Clin._ 71(3), 209–249 (2021). Article PubMed Google Scholar * Luo, G. _et al._ Characteristics and outcomes of pancreatic cancer by histological subtypes. _Pancreas._ 48(6), 817–822
(2019). Article PubMed Google Scholar * Wang, Y. _et al._ Differentiation between non-hypervascular pancreatic neuroendocrine tumors and mass-forming pancreatitis using contrast-enhanced
computed tomography. _Acta Radiol._ 62(2), 190–197 (2021). Article PubMed Google Scholar * Khanna, L. _et al._ Pancreatic neuroendocrine neoplasms: 2020 Update on pathologic and imaging
findings and classification. _Radiographics._ 40(5), 1240–1262 (2020). Article PubMed Google Scholar * Halfdanarson, T. R., Rabe, K. G., Rubin, J. & Petersen, G. M. Pancreatic
neuroendocrine tumors (PNETs): Incidence, prognosis and recent trend toward improved survival. _Ann Oncol._ 19(10), 1727–1733 (2008). Article CAS PubMed PubMed Central Google Scholar *
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. _CA Cancer J. Clin._ 72(1), 7–33 (2022). Article PubMed Google Scholar * Mizrahi, J. D., Surana, R.,
Valle, J. W. & Shroff, R. T. Pancreatic cancer. _Lancet._ 395(10242), 2008–2020 (2020). Article CAS PubMed Google Scholar * Sivapalan, L., Kocher, H. M., Ross-Adams, H. &
Chelala, C. Molecular profiling of ctDNA in pancreatic cancer: Opportunities and challenges for clinical application. _Pancreatology._ 21(2), 363–378 (2021). Article CAS PubMed Google
Scholar * Abbas, M., Alqahtani, M. S., Alshahrani, M. Y. & Alabdullh, K. Aggressive and drug-resistant pancreatic cancer: Challenges and novel treatment approaches. _Discov. Med._
34(173), 158–164 (2022). PubMed Google Scholar * Eissa, M. A. L. _et al._ Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in
blood. _Clin. Epigenetics._ 11(1), 59 (2019). Article PubMed PubMed Central Google Scholar * Follia, L. _et al._ Integrative analysis of novel metabolic subtypes in pancreatic cancer
fosters new prognostic biomarkers. _Front. Oncol._ 9, 115 (2019). Article PubMed PubMed Central Google Scholar * Pereira, S. P. _et al._ Early detection of pancreatic cancer. _Lancet
Gastroenterol. Hepatol._ 5(7), 698–710 (2020). Article PubMed PubMed Central Google Scholar * Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A. & McKusick, V. A. Online
Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. _Nucl. Acids Res._ 33(Database issue), D514–D517 (2005). Article CAS PubMed Google Scholar *
Shoshani, T. _et al._ Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. _Mol. Cell Biol._ 22(7), 2283–2293 (2002). Article CAS PubMed
PubMed Central Google Scholar * Brugarolas, J. _et al._ Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. _Genes Dev._ 18(23),
2893–2904 (2004). Article CAS PubMed PubMed Central Google Scholar * Wang, Z. _et al._ Dexamethasone-induced gene 2 (dig2) is a novel pro-survival stress gene induced rapidly by diverse
apoptotic signals. _J. Biol. Chem._ 278(29), 27053–27058 (2003). Article CAS PubMed Google Scholar * Ellisen, L. W. _et al._ REDD1, a developmentally regulated transcriptional target of
p63 and p53, links p63 to regulation of reactive oxygen species. _Mol. Cell._ 10(5), 995–1005 (2002). Article CAS PubMed Google Scholar * Malagelada, C., Ryu, E. J., Biswas, S. C.,
Jackson-Lewis, V. & Greene, L. A. RTP801 is elevated in Parkinson brain substantia nigral neurons and mediates death in cellular models of Parkinson’s disease by a mechanism involving
mammalian target of rapamycin inactivation. _J. Neurosci._ 26(39), 9996–10005 (2006). Article CAS PubMed PubMed Central Google Scholar * Tirado-Hurtado, I., Fajardo, W. & Pinto, J.
A. DNA damage inducible transcript 4 gene: The switch of the metabolism as potential target in cancer. _Front. Oncol._ 8, 106 (2018). Article PubMed PubMed Central Google Scholar *
Chang, B. _et al._ Overexpression of the recently identified oncogene REDD1 correlates with tumor progression and is an independent unfavorable prognostic factor for ovarian carcinoma.
_Diagn. Pathol._ 13(1), 87 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * Zeng, Q. _et al._ Inhibition of REDD1 sensitizes bladder urothelial carcinoma to paclitaxel by
inhibiting autophagy. _Clin. Cancer Res._ 24(2), 445–459 (2018). Article CAS PubMed Google Scholar * Jin, H. O. _et al._ Redd1 inhibits the invasiveness of non-small cell lung cancer
cells. _Biochem. Biophys. Res. Commun._ 407(3), 507–511 (2011). Article CAS PubMed Google Scholar * Chen, R. _et al._ DNA damage-inducible transcript 4 (DDIT4) mediates
methamphetamine-induced autophagy and apoptosis through mTOR signaling pathway in cardiomyocytes. _Toxicol. Appl. Pharmacol._ 295, 1–11 (2016). Article CAS PubMed Google Scholar * Song,
L. _et al._ DDIT4 overexpression associates with poor prognosis in lung adenocarcinoma. _J. Cancer._ 12(21), 6422–6428 (2021). Article CAS PubMed PubMed Central Google Scholar *
Schwarzer, R. _et al._ REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. _Oncogene._ 24(7), 1138–1149 (2005). Article CAS PubMed Google
Scholar * Du, F. _et al._ DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways. _Cancer Commun. (Lond.)._ 38(1), 45 (2018). PubMed PubMed Central
Google Scholar * Chang, B. _et al._ Overexpression of the recently identified oncogene REDD1 correlates with tumor progression and is an independent unfavorable prognostic factor for
ovarian carcinoma. _Diagn Pathol._ 13(1), 87 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * Di Conza, G. _et al._ The mTOR and PP2A pathways regulate PHD2
phosphorylation to fine-tune HIF1α levels and colorectal cancer cell survival under hypoxia. _Cell Rep._ 18(7), 1699–1712 (2017). Article PubMed PubMed Central Google Scholar * Chang, B.
_et al._ REDD1 is required for RAS-mediated transformation of human ovarian epithelial cells. _Cell Cycle_ 8(5), 780–786 (2009). Article CAS PubMed Google Scholar * Fattahi, F. _et al._
Overexpression of DDIT4 and TPTEP1 are associated with metastasis and advanced stages in colorectal cancer patients: a study utilizing bioinformatics prediction and experimental validation.
_Cancer Cell Int._ 21(1), 303 (2021). Article CAS PubMed PubMed Central Google Scholar * Fattahi, F. _et al._ High expression of DNA damage-inducible transcript 4 (DDIT4) is associated
with advanced pathological features in the patients with colorectal cancer. _Sci. Rep._ 11(1), 13626 (2021). Article ADS CAS PubMed PubMed Central Google Scholar * Feng, Y. _et al._
REDD1 overexpression in oral squamous cell carcinoma may predict poor prognosis and correlates with high microvessel density. _Oncol. Lett._ 19(1), 431–441 (2020). CAS PubMed Google
Scholar * Jia, W. _et al._ REDD1 and p-AKT over-expression may predict poor prognosis in ovarian cancer. _Int. J. Clin. Exp. Pathol._ 7(9), 5940–5949 (2014). CAS PubMed PubMed Central
Google Scholar * Foltyn, M. _et al._ The physiological mTOR complex 1 inhibitor DDIT4 mediates therapy resistance in glioblastoma. _Br. J. Cancer._ 120(5), 481–487 (2019). Article PubMed
PubMed Central Google Scholar * Li, Y. _et al._ ZY0511, a novel, potent and selective LSD1 inhibitor, exhibits anticancer activity against solid tumors via the DDIT4/mTOR pathway. _Cancer
Lett._ 454, 179–190 (2019). Article CAS PubMed Google Scholar * DeYoung, M. P., Horak, P., Sofer, A., Sgroi, D. & Ellisen, L. W. Hypoxia regulates TSC1/2-mTOR signaling and tumor
suppression through REDD1-mediated 14-3-3 shuttling. _Genes Dev._ 22(2), 239–251 (2008). Article CAS PubMed PubMed Central Google Scholar * Ho, K.-H. _et al._ A key role of DNA
damage-inducible transcript 4 (DDIT4) connects autophagy and GLUT3-mediated stemness to desensitize temozolomide efficacy in glioblastomas. _Neurotherapeutics._ 17(3), 1212–1227 (2020).
Article CAS PubMed PubMed Central Google Scholar * Wang, Y. _et al._ Baicalein upregulates DDIT4 expression which mediates mTOR inhibition and growth inhibition in cancer cells. _Cancer
Lett._ 358(2), 170–179 (2015). Article CAS PubMed Google Scholar * Horak, P. _et al._ Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. _Proc.
Natl. Acad. Sci. U.S.A._ 107(10), 4675–4680 (2010). Article ADS CAS PubMed PubMed Central Google Scholar * Kucejova, B. _et al._ Interplay between pVHL and mTORC1 pathways in
clear-cell renal cell carcinoma. _Mol. Cancer Res._ 9(9), 1255–1265 (2011). Article CAS PubMed PubMed Central Google Scholar * Ding, F. _et al._ A review of the mechanism of DDIT4 serve
as a mitochondrial related protein in tumor regulation. _Sci. Progress._ 104(1), 0036850421997273 (2021). Article CAS Google Scholar * Ponting, C. P., Oliver, P. L. & Reik, W.
Evolution and functions of long noncoding RNAs. _Cell._ 136(4), 629–641 (2009). Article CAS PubMed Google Scholar * Martens-Uzunova, E. S. _et al._ Long noncoding RNA in prostate,
bladder, and kidney cancer. _Eur. Urol._ 65(6), 1140–1151 (2014). Article CAS PubMed Google Scholar * Geisler, S. & Coller, J. RNA in unexpected places: Long non-coding RNA functions
in diverse cellular contexts. _Nat. Rev. Mol. Cell Biol._ 14(11), 699–712 (2013). Article CAS PubMed PubMed Central Google Scholar * Arnold, M. _et al._ Global burden of 5 major types
of gastrointestinal cancer. _Gastroenterology_ 159(1), 335–49.e15 (2020). Article PubMed Google Scholar * Hu, J. X. _et al._ Pancreatic cancer: A review of epidemiology, trend, and risk
factors. _World J. Gastroenterol._ 27(27), 4298–4321 (2021). Article PubMed PubMed Central Google Scholar * Aier, I., Semwal, R., Sharma, A. & Varadwaj, P. K. A systematic assessment
of statistics, risk factors, and underlying features involved in pancreatic cancer. _Cancer Epidemiol._ 58, 104–110 (2019). Article PubMed Google Scholar * Ding, F. _et al._ DDIT4 novel
mutations in pancreatic cancer. _Gastroenterol. Res. Pract._ 2021, 6674404 (2021). Article PubMed PubMed Central Google Scholar * Du, F. _et al._ DDIT4 promotes gastric cancer
proliferation and tumorigenesis through the p53 and MAPK pathways. _Cancer Commun. (Lond)._ 38(1), 45 (2018). PubMed PubMed Central Google Scholar * Jin, H. O. _et al._ Induction of HSP27
and HSP70 by constitutive overexpression of Redd1 confers resistance of lung cancer cells to ionizing radiation. _Oncol. Rep._ 41(5), 3119–3126 (2019). CAS PubMed Google Scholar *
Protiva, P. _et al._ Growth inhibition of colon cancer cells by polyisoprenylated benzophenones is associated with induction of the endoplasmic reticulum response. _Int. J. Cancer._ 123(3),
687–694 (2008). Article CAS PubMed Google Scholar * Pinto, J. A., Bravo, L., Chirinos, L. A. & Vigil, C. E. Expression of DDIT4 Is Correlated with NOTCH1 and high molecular risk in
acute myeloid leukemias. _Blood._ 128(22), 5254 (2016). Article Google Scholar * Das, P. K., Islam, F. & Lam, A. K. The roles of cancer stem cells and therapy resistance in colorectal
carcinoma. _Cells._ 9(6), 1392 (2020). Article CAS PubMed PubMed Central Google Scholar * Vinogradov, S. & Wei, X. Cancer stem cells and drug resistance: the potential of
nanomedicine. _Nanomedicine (Lond)._ 7(4), 597–615 (2012). Article CAS PubMed PubMed Central Google Scholar * Lara-Velazquez, M. _et al._ Advances in brain tumor surgery for
glioblastoma in adults. _Brain Sci._ 7(12), 166 (2017). Article PubMed PubMed Central Google Scholar * Kong, D.-S. Cancer stem cells in brain tumors and their lineage hierarchy. _Int. J.
Stem Cells._ 5(1), 12–15 (2012). Article PubMed PubMed Central Google Scholar * Eun, K., Ham, S. W. & Kim, H. Cancer stem cell heterogeneity: Origin and new perspectives on CSC
targeting. _BMB Rep._ 50(3), 117–125 (2017). Article CAS PubMed PubMed Central Google Scholar * Hashemi, F. _et al._ Efficacy of whole cancer stem cell-based vaccines: A systematic
review of preclinical and clinical studies. _Stem Cells._ 41, 207–232 (2022). Article Google Scholar * Du, F. _et al._ DDIT4 promotes gastric cancer proliferation and tumorigenesis through
the p53 and MAPK pathways. _Cancer Commun._ 38(1), 45 (2018). Article Google Scholar * Michel, G. _et al._ Plasma membrane translocation of REDD1 governed by GPCRs contributes to mTORC1
activation. _J. Cell Sci._ 127(Pt 4), 773–787 (2014). CAS PubMed Google Scholar * Lin, L., Stringfield, T. M., Shi, X. & Chen, Y. Arsenite induces a cell stress-response gene, RTP801,
through reactive oxygen species and transcription factors Elk-1 and CCAAT/enhancer-binding protein. _Biochem. J._ 392(Pt 1), 93–102 (2005). Article CAS PubMed PubMed Central Google
Scholar * Wei, C. L. _et al._ A global map of p53 transcription-factor binding sites in the human genome. _Cell._ 124(1), 207–219 (2006). Article CAS PubMed Google Scholar * Chen, K. S.
_et al._ EGF receptor and mTORC1 are novel therapeutic targets in nonseminomatous germ cell tumors. _Mol. Cancer Ther._ 17(5), 1079–1089 (2018). Article CAS PubMed PubMed Central Google
Scholar * Cheng, Z. _et al._ Up-regulation of DDIT4 predicts poor prognosis in acute myeloid leukaemia. _J. Cell Mol. Med._ 24(1), 1067–1075 (2020). Article CAS PubMed Google Scholar *
Qian, C.-N., Mei, Y. & Zhang, J. Cancer metastasis: Issues and challenges. _Chin. J. Cancer._ 36(1), 38 (2017). Article PubMed PubMed Central Google Scholar * Li, J., Wang, Z. &
Li, Y. USP22 nuclear expression is significantly associated with progression and unfavorable clinical outcome in human esophageal squamous cell carcinoma. _J. Cancer Res. Clin. Oncol._
138(8), 1291–1297 (2012). Article CAS PubMed Google Scholar * Liu, X. _et al._ Predictors of distant metastasis on exploration in patients with potentially resectable pancreatic cancer.
_BMC Gastroenterol._ 18(1), 168 (2018). Article ADS CAS PubMed PubMed Central Google Scholar * Daamen, L. A. _et al._ Postoperative surveillance of pancreatic cancer patients. _Eur. J.
Surg. Oncol._ 45(10), 1770–1777 (2019). Article CAS PubMed Google Scholar * Yang, M. & Zhang, C. Y. Diagnostic biomarkers for pancreatic cancer: An update. _World J. Gastroenterol._
27(45), 7862–7865 (2021). Article PubMed PubMed Central Google Scholar * Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: an enhanced web server for large-scale expression
profiling and interactive analysis. _Nucl. Acids Res._ 47(W1), W556–W560 (2019). Article CAS PubMed PubMed Central Google Scholar * Chun, Y. S., Pawlik, T. M. & Vauthey, J. N. 8th
edition of the AJCC cancer staging manual: Pancreas and hepatobiliary cancers. _Ann. Surg. Oncol._ 25(4), 845–847 (2018). Article PubMed Google Scholar * Livak, K. J. & Schmittgen, T.
D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. _Methods._ 25(4), 402–408 (2001). Article CAS PubMed Google Scholar *
Kalantari, E. _et al._ Cytoplasmic expression of DCLK1-S, a novel DCLK1 isoform, is associated with tumor aggressiveness and worse disease-specific survival in colorectal cancer. _Cancer
Biomark._ 33(3), 277–289 (2022). Article CAS PubMed Google Scholar * Rasti, A. _et al._ Co-expression of cancer stem cell markers OCT4 and NANOG predicts poor prognosis in renal cell
carcinomas. _Sci. Rep._ 8(1), 11739 (2018). Article ADS PubMed PubMed Central Google Scholar * Camp, R. L., Charette, L. A. & Rimm, D. L. Validation of tissue microarray technology
in breast carcinoma. _Lab Invest._ 80(12), 1943–1949 (2000). Article CAS PubMed Google Scholar * Jourdan, F. _et al._ Tissue microarray technology: Validation in colorectal carcinoma and
analysis of p53, hMLH1, and hMSH2 immunohistochemical expression. _Virchows Arch._ 443(2), 115–121 (2003). Article CAS PubMed Google Scholar * Shayanfar, N. _et al._ Low expression of
isocitrate dehydrogenase 1 (IDH1) R132H is associated with advanced pathological features in laryngeal squamous cell carcinoma. _J. Cancer Res. Clin. Oncol._ 149(8), 4253–4267 (2022).
Article PubMed Google Scholar * Zlobec, I., Suter, G., Perren, A. & Lugli, A. A next-generation tissue microarray (ngTMA) protocol for biomarker studies. _J. Vis. Exp._ 91, 51893
(2014). Google Scholar * Safaei, S. _et al._ Overexpression of cytoplasmic dynamin 2 is associated with worse outcomes in patients with clear cell renal cell carcinoma. _Cancer Biomarkers._
35, 27–45 (2022). Article CAS PubMed Google Scholar * Kalantari, E. _et al._ Significant co-expression of putative cancer stem cell markers, EpCAM and CD166, correlates with tumor stage
and invasive behavior in colorectal cancer. _World J. Surg. Oncol._ 20(1), 15 (2022). Article PubMed PubMed Central Google Scholar Download references FUNDING This work was supported by
the grant from Iran University of Medical Sciences (Grant Number # 20879). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Oncopathology Research Center, Iran University of Medical Sciences,
Tehran, Iran Fatemeh Tajik, Fahimeh Fattahi, Fereshteh Rezagholizadeh & Zahra Madjd * Department of Surgery, University of California, Irvine, CA, USA Fatemeh Tajik * Clinical Research
Development Unit of Ayatollah-Khansari Hospital, Arak University of Medical Sciences, Arak, Iran Fahimeh Fattahi * Department of Molecular Medicine, Faculty of Advanced Technologies in
Medicine, Iran University of Medical Sciences, Tehran, Iran Fereshteh Rezagholizadeh & Zahra Madjd * Department of Pathology, School of Medicine, Iran University of Medical Sciences,
Tehran, Iran Behnaz Bouzari, Pegah Babaheidarian & Zahra Madjd * Department of Surgery, Firoozgar Hospital, Iran University of Medical Sciences, Tehran, Iran Masoud Baghai Wadji Authors
* Fatemeh Tajik View author publications You can also search for this author inPubMed Google Scholar * Fahimeh Fattahi View author publications You can also search for this author inPubMed
Google Scholar * Fereshteh Rezagholizadeh View author publications You can also search for this author inPubMed Google Scholar * Behnaz Bouzari View author publications You can also search
for this author inPubMed Google Scholar * Pegah Babaheidarian View author publications You can also search for this author inPubMed Google Scholar * Masoud Baghai Wadji View author
publications You can also search for this author inPubMed Google Scholar * Zahra Madjd View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Z.M.
designed and supervised the project, rechecked and approved all parts of the manuscript, and conducted data analysis. F.T. wrote the manuscript, performed the IHC experiment, and analyzed
and interpreted the SPSS data. F.F. analyzed the RT-qPCR data, interpreted the bioinformatics data, wrote the bioinformatics parts of the manuscript, and helped to prepare the RT-qPCR
figures. F.T. and F.R. collected the fresh tissue and paraffin-embedded tissues, collected the patients’ clinicopathological data and survival data, and also performed an RT-qPCR experiment.
B.B. marked the most representative areas in different parts of the tumor for the construction of TMA blocks. B.B. and P.B. scored TMA slides after IHC staining and helped to prepare the
IHC figures. M.B.W. was the surgery specialist who helped collect the fresh samples and access the patient’s data. All authors read and approved the final manuscript. CORRESPONDING AUTHOR
Correspondence to Zahra Madjd. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the
article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your
intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence,
visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Tajik, F., Fattahi, F., Rezagholizadeh, F. _et al._ Nuclear overexpression of
DNA damage-inducible transcript 4 (DDIT4) is associated with aggressive tumor behavior in patients with pancreatic tumors. _Sci Rep_ 13, 19403 (2023).
https://doi.org/10.1038/s41598-023-46484-3 Download citation * Received: 08 April 2023 * Accepted: 01 November 2023 * Published: 08 November 2023 * DOI:
https://doi.org/10.1038/s41598-023-46484-3 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
