Identifying symptom etiologies using syntactic patterns and large language models
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Differential diagnosis is a crucial aspect of medical practice, as it guides clinicians to accurate diagnoses and effective treatment plans. Traditional resources, such as medical
books and services like UpToDate, are constrained by manual curation, potentially missing out on novel or less common findings. This paper introduces and analyzes two novel methods to mine
etiologies from scientific literature. The first method employs a traditional Natural Language Processing (NLP) approach based on syntactic patterns. By using a novel application of
human-guided pattern bootstrapping patterns are derived quickly, and symptom etiologies are extracted with significant coverage. The second method utilizes generative models, specifically
GPT-4, coupled with a fact verification pipeline, marking a pioneering application of generative techniques in etiology extraction. Analyzing this second method shows that while it is highly
precise, it offers lesser coverage compared to the syntactic approach. Importantly, combining both methodologies yields synergistic outcomes, enhancing the depth and reliability of etiology
mining. SIMILAR CONTENT BEING VIEWED BY OTHERS VALIDATING LARGE LANGUAGE MODELS AGAINST MANUAL INFORMATION EXTRACTION FROM CASE REPORTS OF DRUG-INDUCED PARKINSONISM IN PATIENTS WITH
SCHIZOPHRENIA SPECTRUM AND MOOD DISORDERS: A PROOF OF CONCEPT STUDY Article Open access 20 March 2025 NATURAL LANGUAGE PROCESSING ANALYSIS OF THE THEORIES OF PEOPLE WITH MULTIPLE SCLEROSIS
ABOUT CAUSES OF THEIR DISEASE Article Open access 24 June 2024 A PHENOTYPE-BASED AI PIPELINE OUTPERFORMS HUMAN EXPERTS IN DIFFERENTIALLY DIAGNOSING RARE DISEASES USING EHRS Article Open
access 28 January 2025 INTRODUCTION Differential diagnosis is a cornerstone of modern medicine, holding paramount importance in the field. It serves as a systematic and meticulous process
that allows healthcare professionals to distinguish between various potential medical conditions with similar symptoms. By considering a range of possible diagnoses and eliminating them one
by one, clinicians can arrive at an accurate diagnosis, ensuring that patients receive appropriate treatment and care. This critical step not only enhances patient outcomes but also
minimizes the risks of misdiagnosis and unnecessary procedures, highlighting the indispensable role of differential diagnosis in the practice of medicine. Clinicians often consult resources
such as books, review papers, or summarization services like UpToDate1 in order to understand possible etiologies for a set of symptoms. However, these resources may be limited in their
ability to incorporate the latest findings or less common ones, as they are manually curated. Automated text mining has been proposed as a solution to address these limitations, but text
mining efforts have typically focused on small subsets of data or required NLP (Natural Language Processing) experts to operate complex pipelines2,3,4,5,6,7. As a result, these efforts have
not led to the creation of up-to-date resources widely used by the scientific community. In this study, we suggest two methods for mining etiologies from the scientific literature. The first
method is rooted in traditional NLP and makes use of syntactic patterns and iterative bootstrapping. In contrast to previous studies in pattern bootstrapping which relied on fully automated
pattern creation2,8,9, we advocate a human-in-the-loop approach. We developed a dedicated bootstrapping user interface (UI) and showed that by using it, a set of syntactic patterns could be
obtained with a reasonable human effort (4 h) and that the patterns could be used to reliably extract symptom etiologies with high coverage. The second method makes use of the ChatGPT-4
(Chat-Generative Pre-trained Transformer-4) generative model to generate etiologies and employs a fact verification pipeline to remove hallucinations and provide reliable provenance
information. This is to our knowledge the first systematic application of generative methods to etiology extraction. We showed that this method can be highly accurate, but has lower coverage
compared to syntactic patterns running on a large corpus. This is consistent with recent findings about the difficulty of LLMs (Large Language Models) in encoding the long tail of world
knowledge10,11. Importantly, as we’ll show in the “Results” section, the method of syntactic patterns and the generative method complement each other, and the best overall results are
obtained by combining both. An important feature of this study is that the relevant extraction patterns and GPT (Chat-Generative Pre-trained Transformer) prompts may be applied to extract
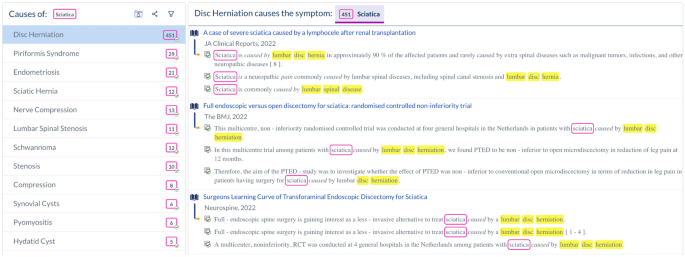
symptom etiologies from live and up-to-date resources, at interactive speed. While the process is described in detail in this paper, we also provide a webpage which abstracts the details and
allows researchers and clinicians to search for symptom etiologies using these methods and based on symptom name alone. Figure 1 shows a screenshot of this page, where etiologies for
sciatica are listed, along with their provenance information. The contributions of this study can be summarized as follows. * 1. A workflow and dedicated UI to bootstrap syntactic patterns.
The workflow has been applied in this study to identify patterns for etiologies, but could easily be applied to other relations between entities. * 2. A novel application of ChatGPT with a
dedicated fact verification pipeline to the problem of identifying etiologies. * 3. A dedicated UI which allows to easily retrieve etiologies from a live and routinely updated snapshot of
the scientific literature. * 4. Analysis of the current state of text mining for etiology detection, and a comparison to alternative resources such as books, review papers and UpToDate).
RELATED WORK There has been a significant amount of research on extracting information from the scientific literature, particularly in the biomedical domain12,13. Many approaches have
focused on extracting common biomedical entities like genes, diseases and drugs and the relationships between them. However, less work has been directed at extracting symptom etiologies.
Extracting etiologies is a critical task for commercial symptom checkers such as WebMD14, patient.info15 and Isabel16 which apply computation inference on top of pre-curated symptom-disease
data. The data required for these systems is acquired from medical books and top journals and is curated manually by medical experts17. The data is accurate but requires domain expertise and
vast amounts of time to curate and update. Furthermore, rare or very recently discovered etiologies are often overlooked and the curated etiologies lack fine-grained frequency information.
An alternative approach for disease-symptom extraction is based on data mining and co-occurrence. This involves extracting diseases and symptoms from medical sources, normalizing the
extracted names based on medical vocabularies (relevant vocabularies are DO, SYMP, UMLS, MeSH, MEDIC, SNOMED-CT) and using disease-symptom co-occurrence (e.g. co-occurrence in the same
PubMed abstract, or the same EMR report) to statistically model the disease-symptom connections. This approach is typically used in the construction of disease networks, where the extracted
disease-symptoms associations are used to populate a knowledge graph, often along with other types of connections like disease-gene or disease protein7,18,19,20. Disease networks include
fine-grained frequency information and are useful for statistical inference, discovering connections between seemingly unrelated diseases and for drug repurposing. However, this approach
does not directly extract causal disease-symptom relations, and the reliance on co-occurrence introduces a degree of noise which makes it less relevant for a clinical setting where
clinicians require etiologies to be accurate and/or easily validated by accompanying evidence. More similar to our approach are studies using syntactic patterns over dependency trees to
extract disease-symptom relations2,4. For example, Hassan et al.4 manually annotated a dataset of labelled relations (PubMed sentences with at least diseases and symptom spans, and a set of
zero or more binary relations between disease-symptom pairs) and used it to generate syntactic patterns. The generated patterns were then ranked and selected based on quality criteria such
as the ratio of the correct pattern extractions to the total extractions. Our work differs from these approaches by not relying on labelled data and instead employing iterative bootstrapping
and a dedicated UI facilitating a human-in-the-loop approach to pattern creation (see section “Results”/“Syntactic patterns and iterative bootstrapping”). In addition, these earlier works
did not release models, training data or extractions, so their outputs are not available to medical professionals. Concerning Machine Learning (ML) modelling, Disease Named Entity
Recognition (NER) datasets have existed for a while21,22, but a dataset for symptom extraction was only recently released23. Furthermore, Feng et al.3 are as far as we know the only ones
applying ML techniques to disease-symptom extraction. This is most likely due to the absence of sizable training data for this problem. In their study Feng et al. trained kernel support
vector machines (SVMs) for relation disease-symptom relation classification in a semi-supervised setting. (In part of this work they did hand label a small ad-hoc dataset to train and
evaluate their model, but their main contribution was at developing models that can utilize unlabeled data). Models, code or extractions were not made available as part of this work. RESULTS
Below we briefly describe the suggested methods for etiology extraction and evaluate their results. SYNTACTIC PATTERNS AND ITERATIVE BOOTSTRAPPING Figure 2 shows the general architecture of
our approach, comprising three main stages: (1) bootstrapping: a dedicated UI component allowing pattern developers to quickly identify syntactic extraction templates based on a handful of
result examples. The UI component has been made publicly available as part of the SPIKE extractive search system. The SPIKE system is a free online information extraction platform available
at https://spike.apps.allenai.org. As part of this work we have added to it a module for bootstrapping syntactic patterns which is described in the “Methods” section; (2) extraction: the
system applies the patterns, removes duplicates from the results and obtains sentences with etiology mentions; and (3) mention unification: the extracted mentions are unified into groups of
synonymous mentions (“concepts”). The concepts are then ranked by their number of mentions. In the methods section we describe each of these stages in more detail. GENERATING ETIOLOGIES
USING CHATGPT The second method we suggest makes use of generative models, specifically ChatGPT. Since generative models suffer from hallucinations, we employ a novel fact verification
pipeline with an evidence ranking component to verify the generated etiologies and provide provenance information. Figure 3 describes the process which is comprised of 3 stages: (a) a
generative model is prompted to generate an exhaustive list of symptom etiologies; (b) For each generated etiology, we use dense retrieval to obtain N candidate evidence sentences; (c) for
each etiology and N candidate evidence sentences, we task a generative model with selecting and ranking n ≪ N sentences which would constitute the best supporting evidence for the etiology.
We ask that sentences where the causality is expressed explicitly will be ranked above sentence where it is expressed implicitly, and that sentences that do not express or imply causality
between an etiology and the symptom are removed; and (d) we use a final prompt to the language model to verify that the best selected evidence is indeed valid evidence. The “Methods” section
describes each of these stages in detail. EVALUATION In evaluating the two suggested approaches, we look to measure their recall—the percentage of etiologies they identify out of the full
set of etiologies for a symptom—and precision—how many of the etiologies they identify are indeed correct. While estimating the precision of the approaches is possible through sampling,
robustly estimating their recall turns out to be challenging since as we’ll see below, reference sources are themselves very incomplete, and so there is no reliable source for the full set
etiologies for a symptom. To mitigate this problem, we borrow an evaluation approach used in information retrieval, where etiologies are pooled from several different sources (for each
symptom, we use our two suggested approaches to identify etiologies plus a reference source with etiologies known to be correct), results are then manually labelled by experts, and we
consider the full set of correctly identified etiologies to be our full set. The recall figure for the different systems is then taken as the percentage of extractions they identify from
this full set. This method requires normalizing the extractions of the different systems so that an etiology which appears with different names in the different sources is indeed treated as
a single etiology. This method of evaluation is detailed in the section “Comprehensive evaluation with reference sources” and it allows us to estimate the recall of the different approaches
and point out their limitations. However, due to the large number of etiologies extracted for each system, and the need for name normalization, this approach does not scale easily to a large
number of symptoms. To further evaluate the precision of our approaches across many symptoms, we randomly select a larger number of symptoms and then several random extractions for each
symptom. Annotators then labelled the correctness of these random extractions. The results are detailed in the section “Evaluating precision via random sampling” and provide further evidence
for the precision of the two approaches at a larger scale. COMPREHENSIVE EVALUATION WITH REFERENCE SOURCES For this experiment, we extracted etiologies for 3 symptoms: hiccups, jaundice and
chest pain. For each of these symptoms, we obtained a list of etiologies from a reference source: for hiccups, we accessed the table “Causes of persistent and intractable hiccups”24 in
UpToDate on 01/12/2022, for Jaundice we used a book source25. For chest pain, we also used an UpToDate summary 26 accessed on 01/12/2022. Since both the syntactic patterns and GPT provide a
substantial number of supported etiologies which do not appear in the reference source, we first pool together the etiologies from all sources (the patterns, GPT and the reference),
normalize the names manually so that the terminology is the same across sources, and then have a team of trained annotators validate the extractions (we employed a team of 3 annotators, all
4’th year medical students in the Technion medical school, Israel. Annotators were paid for their participation in the annotation effort). Etiologies from the reference source were assumed
to be supported by evidence. Etiologies obtained from the Patterns or GPT (and not found in the reference) were labelled as supported/unsupported (correct/incorrect) based on the provided
provenance information. both the patterns and the GPT’s verification pipeline provide provenance information in the form of sentences which mention the symptom and the candidate’s etiology
and provide information supporting the causal relation between them. The annotators additionally evaluated the correctness of the unvalidated extractions of ChatGPT, in these cases, they
were instructed to rely on external sources (namely Google and Pubmed searches) for their annotation. The annotation effort was supervised by an expert MD (Dr Maayan Ben Sasson). We first
measured agreement between the expert and each of the annotators on 20 random extractions (10 from the GPT pool and 10 from the patterns pool) and noted high agreement in all cases.
Cohen's kappa agreement was 1 between the expert and the Jaundice annotator, 0.86 between the expert and the Hiccups annotator and 0.77 between the expert and the Chest Pain annotator.
Each of the annotators then proceeded to annotate the rest of the extractions for their respective symptoms. Figure 4 includes Venn diagrams depicting the overlap between the supported
etiologies provided by the 3 different sources. As shown in the figure, the patterns have better overall coverage, typically identifying many supported etiologies which are not listed in the
reference or identified by ChatGPT. Importantly, despite a degree of overlap, the patterns and GPT have a significant number of supported and non-overlapping extractions, suggesting that it
may be useful to combine the two methods. Table 1 shows the full results for the different systems, taking into account unsupported (incorrect) extractions. The first two rows in Table 1
show the results for the patterns and GPT systems. As expected from the Venn diagrams, the patterns have a substantially higher recall than GPT. This is following recent observations about
the difficulty of generative models to handle the “long tail” of corpus facts10,11. However, GPT has significantly higher overall precision, even without applying its fact verification
pipeline. In the third row of Table 1, we show the results of applying the fact verification pipeline, which brings GPT’s precision to 100% on all symptoms, at the cost of somewhat lower
recall. The final two rows of Table 1 show the results of aggregating the pattern extractions and GPT generations. Unifying the non-verified variant of GPT with the patterns yields the
overall highest recall and F-Score compared to all other solutions while unifying the patterns with the verified variant of GPT (labelled GPTV in the table) provides slightly lower Recall
and F-Score, but higher precision. Both aggregated variants provide significantly better results than using GPT or the patterns by themselves. Table 2 shows the recall of the different
solution when evaluated only against the reference sources, without considering the full spectrum of verified etiologies. The syntactic patterns still show the highest recall of the
non-aggregated methods, and the combination of the patterns and GPT still shows the highest overall recall. However, when evaluating against the reference sources alone, the difference in
coverage between the patterns and GPT is much less significant. This is in line with the observation that LLMs are effective at encoding the more common knowledge, which is likely to appear
in curated reference sources, and less effective at encoding the long tail knowledge11. Table 3 shows a sample of the extractions of the different systems, the full set of extractions and
annotations is available in Supplementary Materials 1. EVALUATING PRECISION VIA RANDOM SAMPLING In this experiment, we sampled 20 symptoms from the CTD list of signs and symptoms.
(https://ctdbase.org/downloads/;jsessionid=38DDC4627CA9AC7E6EE1120C8284E879#alldiseases.) We then sampled 10 etiologies for each symptom: 5 were sampled randomly from the GPT generations and
5 from the pattern extractions. 2 annotators (An expert MD and a cancer researcher, both authors of this paper) labelled the extractions as 0 (incorrect) and 1 (correct). We measured
agreement on 30 extractions before labelling the full set (Cohen’s kappa = 0.71). Table 4 summarizes the results and we include the full annotations in Supplementary Materials 2. The results
across all etiologies show 91% precision for ChatGPT and 81% precision for the patterns approach. These results are in line with the precision metrics measured in the comprehensive
evaluation of the 3 symptoms (Table 1) where macro-averaged GPT results were slightly higher (94.6%) and the pattern results were slightly lower (75.3%). In the Qualitative Evaluation
section, we’ll review some of the mistakes made by both approaches. In Table 4 we also list the yield of the two approaches (number of generated/extracted etiologies for each symptom). While
the numbers may be slightly exaggerated due to the lack of manual name normalization, they do further attest to the high number of etiologies identified, especially by the patterns
approach. QUALITATIVE ANALYSIS To better understand the mistakes made by the two approaches we looked at etiologies missed by each approach (false negatives) and etiologies which were
incorrectly identified (false positives). We’ll address the two cases below. MISSED ETIOLOGIES A known limitation of GPT and LLMs is that they can not encode the long tail of rare facts in
its parameters10,11, so in considering missed etiologies we focused on etiologies listed in our reference sources but not identified by our patterns approach, which has generally high
recall. We sampled 20 such etiologies and tracked down the cited sources for these etiologies in the reference. We then mapped out the reasons these sources were not identified by the
patterns. The results are summarized in Table 5 below: The data in the table demonstrates a few limitations of the system. First, for 6 etiologies listed as “Coludn’t validate source,” we
were unable to track the relevant cited text. This is either because the cited text is not publicly accessible, or in some cases, the cited text did not mention the etiology. This highlights
a limitation of the extractive patterns approach, namely that not all relevant medical information is in the form of digital and publicly accessible publications. This issue cannot be
readily addressed, but we’re hopeful that the situation will gradually improve as more publications transition into open-access modes. Another notable problem which occurred 9 times (the
aggregate of “Tabular Data, Not in Abstract” and “Not in Abstract”), is that the relevant information did not appear in the abstracts of PubMed paper, but rather in the body text (either in
text form or as tabular data). This should be partially addressed by future updates to the SPIKE system which is expected to cover full texts of ~ 4 million PubMed papers (the PMC
collection) in addition to its current coverage of ~ 25 million abstracts. Tabular data is not currently supported in the SPIKE system so this will remain a limitation for the time being and
may be addressed by future research. 4 of the missing etiologies were cases where the relevant information appeared in a PubMed abstract, but was not captured by our patterns. A case in
point was the “hiccups” symptom, where certain drugs were listed in the reference source as etiologies and our patterns did not identify them. Indeed, patterns for drug side effects were not
added as part of our initial bootstrapping effort as these were not relevant for the symptoms considered there. We plan to add additional patterns to address this shortcoming. However, it
should be noted that there’s a long tail of ways researchers can indicate causation between an etiology and a symptom, so pattern coverage will always remain incomplete to some degree.
INCORRECTLY IDENTIFIED ETIOLOGIES To understand the reasons for incorrectly identified etiologies in our two approaches we sampled 10 false positives predicted by each method. Table 6
summarizes the types of incorrect etiologies generated by GPT. The main source of errors were cases where the generated etiology is related to a correct etiology but is not in itself
accurate. Another class of errors are cases where GPT generated correct causes, but these were unlikely to be considered useful etiologies by medical professionals (e.g. due to being too
generic and non-specific). Table 7 summarizes the types of errors made by the patterns approach. The errors made by pattern application are different from the ones made by GPT and stem from
errors made by the different NLP components in the extraction pipeline (e.g. incorrect expansion of a word to its linguistic phrase, parsing errors made by the parser) or by patterns which
are not restrictive enough and generate some noise in addition to useful results. METHODS EXTRACTING ETIOLOGIES USING PATTERNS EXTRACTIVE SEARCH FOR ITERATIVE BOOTSTRAPPING We implemented
the suggested human-in-the-loop variant of iterative bootstrapping as part of the SPIKE-PubMed extractive search system. In extractive search systems27,28,29, users issue queries which
consist of a sequence of terms, some of which are designated as capture slots. The capture slots act as variables, whose values should be extracted (“captured”) from matched sentences. For
example, the query in (1) is a structured ES query which retrieves all sentences which contain the word “caused”, connected by an edge of _nsubpass_ (passive subject) to a word which will be
designated as symptom and edge of _nmod_ (nominal modifier) to a third word which will be designated as a disease. All PubMed sentences are pre-parsed and indexed and so the syntactic
relations between words are being enforced by the matcher. Figure 5 below shows the query graph which corresponds to the structured query. * (1) STRUCTURED < > :sciatica is $caused by
< > :endometriosis The dollar preceding the word “caused” indicates that this word should be matched as-is. The colon preceding the terms _sciatica_ and _endometriosis_ terms
indicates that these are captured slots. This means that the matching sentences are not required to include these precise words. Rather they are required to include words that are in a
similar syntactic configuration to these terms (i.e. connected by _nsubjpass_ and _nmod_ edges respectively to the word “caused”, respectively). These captured words are then extracted,
expanded to their linguistic phrase boundaries, aggregated and presented to the user as symptom-etiology pairs. In addition to structured queries that match terms based on their syntactic
relationships, extractive search systems provide a “basic” query type, that can match terms irrespective of their position in a sentence as well as other types of queries. The common
property of all queries is the ability to designate query terms as capture slots, triggering span extraction in addition to sentence matching. The support for both syntactic queries and
basic/keyword queries lends itself well to implementing a workflow for iterative bootstrapping, as described below in the section “Methods”/“Generating syntactic patterns using iterative
bootstrapping”. GENERATING SYNTACTIC PATTERNS USING ITERATIVE BOOTSTRAPPING As part of this study, we extended the SPIKE-PubMed extractive search system with a module for iterative
bootstrapping (it is now integrated into the SPIKE engine as part of its “Suggestions” section). Using this module, pattern developers start by defining one or more seed tuples expressing
the relation of interest (e.g. <sciatica, herniation> was the only initial seed tuple used in generating patterns for the symptom-etiology relation). The system then searches PubMed
for sentences that contain both tuple elements of one of the seed tuples. The system then automatically generates syntactic patterns from each matching sentence based on the syntactic path
between the two matched elements (see “Methods”/“Generating patterns from matching sentences”). Each pattern has a user-friendly text representation and the patterns are ranked by yield. The
pattern developer can quickly inspect the yield of each pattern and decide whether to accept or reject it. The system then uses the selected patterns to extract more seed tuples and the
process is repeated iteratively. The pattern developer can choose to stop the process once the newly discovered patterns become very specific as indicated by their low yield (e.g. when a
pattern matches only the sentence from which it was generated, or up to N additional sentences). Figure 6 shows a screenshot of the initial set of patterns suggested by the bootstrapping
component based on the <sciatica, herniation> seed tuple. In parenthesis, before each pattern, we see the number of matched sentences which triggered it. The pattern developer can
click the search button (the magnifying glass) to inspect the sentences, and press the plus icon to add a pattern to the pattern pool. The UI was used by a medical information specialist to
derive extraction patterns for the symptom-etiology _caused by_ relation in the following manner: * 1. Starting from a single seed <sciatica, herniation>, the system retrieved all
PubMed sentences which contained the words _herniation_ and _sciatica_ * 2. The system generated patterns from all matching sentences, and the information specialist inspected all patterns
with a yield greater or equal to 2. 16 initial patterns were selected and added to the pattern pool. (Note that at this stage, the information specialist quickly inspected the pattern yield,
and eliminated patterns with low precision. For example a pattern like “sciatica with _____” was suggested as it’s common to find etiologies in the position of the placeholder (e.g.
“sciatica with disk herniation”), however, the pattern was discarded by the information specialist as it does not imply causation and frequently extracts results like “symptoms included
sciatica with pain and numbness” where pain and numbness are not etiologies.) * 3. The system then ran the selected patterns yielding new symptoms and etiologies. We collected the top 10
symptoms and top 50 etiologies for another round of iterative bootstrapping. * 4. We re-ran steps 1–3 with the new seeds, again inspecting patterns with yield greater or equal to 2. This
resulted in an additional 44 patterns reaching a total of 60 patterns. Re-applying the process did not result in new high-yield patterns so we stopped after two iterations. The full set of
patterns is given in Supplementary Materials 3. GENERATING PATTERNS FROM MATCHING SENTENCES The input to the pattern generation process is a sentence _S_ and a pair of tuple elements in S,
denoted with A and B (e.g. the sentence S may be “Sciatica is caused by disk herniation” and the matched tuple elements are A = ” Sciatica” and B = ” disk herniation”). We’ll denote with T
the universal (basic) dependency tree for S. The system derives a pattern from this input in the following way: * (1) Obtain the shortest undirected syntactic dependency path _P_ = _[A, n1,
n2, …, B]_ between _A_ and _B_ in _T_. * (2) If P includes only A and B (i.e. the two items are directly connected in T), find C which is the lowest mutual ancestor of A and B. If C exists,
set P to be the path from A to C to B. * (3) Derive a linearized pattern from P by walking through the items on the path, including the dependency edges between pairs of items and the
lemmatized form of the words along the path. For example, for the sentence “[sciatica] in low back pain is usually caused by disc [herniation]” and the tree in Fig. 7, the generated path
from “sciatica” to “herniation” is: > _[symptom placeholder] :_ <_nsubjpass : lemm_=_cause :_ >_nmod_by : > [disease placeholder]_ The pattern will match sentences where a word
with the lemma cause is connected by an outgoing _nsubjpass_ edge to a word representing the symptom and by an outgoing _nmod_by_ edge to a word representing an etiology. The matched words
for symptom and etiology are automatically expanded by the SPIKE system to their linguistic phrase boundaries (e.g. if the word “herniation” is matched in the etiology position, it may be
expanded to “disc herniation” or “lumbar disc herniation” depending on its surrounding context). Note that superfluous elements not on the path such as “in low back pain”, “is usually” and
“by disc” are dropped from the path so the resulting syntactic pattern will generalize better than a sequence pattern derived from the same sentence. PATTERN PROCESSING Once a set of
patterns is obtained, we use the SPIKE runtime to apply the patterns over PubMed. We obtain matching sentences and obtain sentences and pairs of symptom-etiology pairs in them which satisfy
the syntactic restrictions. This results in a set of “Etiology Mentions”: sentences with marked etiology spans. GROUPING MENTIONS INTO CONCEPTS To group mentions into concepts we proceed
with normalization and abbreviation detection: * Normalization: we take from each raw mention its string fingerprint based on the steps below. We then unify all the mentions with the same
fingerprint into a single concept. * We lowercase the name, remove punctuation, remove prefixes (a/an/the etc.), remove preceding/terminating spaces and turn multiple spaces into a single
space * Split tokens on whitespace and remove duplicates * Sort the tokens to normalize word orders * If the resulting normalized name is greater than 6 chars, we run a Soundex algorithm
(shorter strings remain as is). * Abbreviation detection: we detect abbreviations using the Hearst Algorithm30 and replace them with their expanded form. * Linking: we use the BioSyn entity
linker31 to link the entities into the MEDIC disease ontology32. We take the top result of BioSyn (i.e. the entry name in MEDIC which is most likely to be associated with the entity)
irrespective of its score, and use it as a concept ID. Since BioSyn always returns linking targets, the mentions will be given IDs even if a corresponding concept does not exist in MEDIC.
However, since the IDs are only used for grouping it’s uncommon for this to pose a problem, as such entries will not typically be grouped with other entries, and will remain as singletons,
maintaining their original (correct) name. GENERATING ETIOLOGIES USING CHATGPT We used the May 12th, 2023 version of ChatGPT 4.0 (and in the ablations involving GPT 3.5, we used the version
from the same date as GPT 3.5). ChatGPT has been used for Item Generation, Evidence Ranking and Evidence Selection, for Evidence Retrieval we used the SPIKE extractive search engine with a
PubMed index which contains all PubMed abstracts published before February 2023. We experimented with different prompts and parameter options for the different stages. In all experiments,
ChatGPT was run through the OpenAI API with temperature = 1, max_tokens = 4096 and top_p = 1. Below we specify the final prompts selected for each stage as well as some of the alternatives
considered. ITEM GENERATION To generate a list of items we used the prompt below, where <symptom> is replaced with one of our target symptoms (hiccups, jaundice or chest pain). >
“Provide an exhaustive list of all possible <symptom> etiologies. > Be succinct, list only names, each name in its own line. Include all > known etiologies, even the most rare
ones. write done when you > finish listing all etiologies.” To obtain the prompt we had 3 medical professionals spend 30 min trying out alternative prompts for symptoms they were familiar
with (but not the ones tested) and send us the one that seemed best to them. We then compared the results of the 3 prompts on a target symptom (sciatica) and chose the best one. For each
test symptom, we then ran the prompt 3 times and selected the run with the most results. Note that there is a difference in the number of results generated by GPT for the same symptom across
runs (up to 17 fewer generated etiologies across symptoms in our results). Merging the results of multiple runs is non-trivial since GPT sometimes refers to the same etiology in slightly
different names between runs. Still, if the issue of normalization is addressed, we believe this is an interesting direction for improving the recall of GPT. Also, ChatGPT can be asked to
add more results to its initial result list. We experimented with this but found that it greatly increases hallucinations and variance. EVIDENCE RETRIEVAL To retrieve evidence for a specific
candidate etiology we used the dense retrieval options of the SPIKE engine based on the abstract embeddings described in Ravfogel et al. 33. In this case, SPIKE relies on a pre-populated
embedding space which includes embeddings for all PubMed sentences. When given a query sentence, SPIKE retrieves the sentences which are semantically closest to it in vector space. The
embeddings in Ravfogel et al. 33 are especially suited for cases where the search query contains an abstract term (e.g. “etiology”) and the matched sentences contain specific instances of
the abstract term (e.g. specific diseases, etc.). We used the query “an etiology for <symptom>” to find sentences which express a causal relation between an etiology and the symptom at
hand. While similarity search is by design soft, we also added hard constraints requiring the results to contain the exact phrases for the etiology and symptom. We retrieved the top 50
matching sentences for each etiology. Note that adding the hard constraints does mean we sometimes lose useful results (i.e. PubMed sentences where the symptom or etiology are mentioned in a
different name than the one suggested by GPT). This is one of the causes of reduced recall by the “verified” GPT variant. However, as shown in the ablations in Table 8 below, without these
hard constraints the number of etiologies with retrieved sentences increases (as expected, as the search is less stringent), but the retrieved sentences are less relevant, so after ranking
and vetting the candidate evidence, the overall number of valid results for which evidence is found is decreased significantly. EVIDENCE RANKING To rank the evidence we used the prompt: >
“From the list of sentences below (one per line), which 3 > sentences would be the most convincing ones to be used as evidence > that <candidate_etiology> is an etiology for
<symptom>? > > When selecting the 3 sentences, prioritize sentences which > explicitly state that the item belongs to the list over ones where > this is only implied. >
> Only select sentences from the list below, do not add new sentences. > If the list does not contain 3 sentences that could be used as > supportive evidence, return fewer
sentences, or 0 sentences (an > empty list) if needed. > > Format the results as a Python array, where each item is a dict with > two items, the first item ("sent"),
is the sentence itself. The > second item ("reason") is a textual explanation of the reason why > the sentence was selected. > > Be succinct, reply with the Python
list and nothing else.” As evidence ranking is applied once per retrieved etiology it is potentially expensive to run at scale. We tested the effects of using GPT 3.5 turbo which is a
magnitude of order cheaper than GPT 4.0. The results in Table 9 indicate a significant degradation when GPT 3.5 is used instead of GPT 4.0. EVIDENCE VETTING For evidence vetting, we used the
following prompt. > “Can the following sentence be used as evidence that "<candidate > etiology>" is an etiology for <symptom>? Be succinct, and reply with >
yes or no only. > > The sentence is: > > <evidence_sentence>” As evidence vetting can potentially be expensive at scale, we tested the effects of using GPT 3.5 turbo
which is a magnitude of order cheaper than GPT 4.0. The results are listed in Table 10. Compared to evidence ranking, where the use of GPT 4.0 appears critical, results of evidence vetting
were less clearly impacted by the use of GPT 3.5 (2 symptoms showed better and slightly better results with GPT 4, while the third symptom showed slightly better results with GPT 3.5).
DISCUSSION In this study, we presented two novel methods to address the important medical problem of extracting symptom etiologies from the scientific literature. We show that generative
models (ChatGPT) provide accurate results, especially when combined with the suggested verification pipeline, however, they struggle with retrieving the long tail of relevant etiologies.
Patterns on the other hand have lower accuracy, but since they’re applied to an index of all Pubmed abstracts they provide much higher recall, compared both to GPT and the reference sources.
Importantly, as shown in the Venn diagrams in Fig. 4 and the results in Table 1, both solutions turn out to be complementary and the best results are obtained by combining them. Below we
discuss additional interesting aspects of this work. THE PROBLEM OF INCOMPLETE COVERAGE An interesting observation of this study is that even established manually curated clinical sources
such as UpToDate or medical books are highly incomplete in their coverage of relevant symptom etiologies. Generative methods are also not ideal at covering the long tail of etiology data, as
they struggle to encode rare facts in their limited parameter space11. Patterns applied over a large corpus (in this case the set of all PubMed abstracts) are still the most effective
individual method at identifying the long tail of relevant information, but these also suffer from partial recall, not extracting the full set of etiologies identified by the other sources,
and thus cannot fully replace them. In the “Results”/“Qualitative analysis” section, we discuss the issues preventing the patterns from achieving higher recall, pointing out legal
restrictions and data availability as significant issues, but also suggesting actionable steps which may improve recall with available data, namely applying the patterns to a larger part of
the open scientific data (through the use of PMC-PubMed, in addition to PubMed abstracts), and addressing issues of missing patterns and findings organized in Tabular form. The application
of generative methods to this problem may also benefit from recent advancements in retrieval augmented language models34 and systematically applying such models to this problem may be a
promising area for future research. RELEVANCE TO MEDICAL PROFESSIONALS Louis Pasteur's famous quote: “In the fields of observation, chance favours only the prepared mind" means
that the better prepared and more knowledgeable you are, the more you'll be able to take advantage of any chance opportunities or observations. Most of the landmark studies that have
changed clinical medicine have been published in journals. Over the years, the number of active, peer-reviewed journals has expanded to approximately 28,000, collectively publishing more
than 1.8 million articles every year35. In such an era, a ``prepared mind`` needs a tool to process such an amount of knowledge. We demonstrated the capacity to process vast volumes of
medical literature and healthcare data in real time, thereby granting medical professionals access to the most current and comprehensive information available. This stands in stark contrast
to manual searches, which often yield outdated or incomplete results, potentially culminating in misdiagnosis or delayed treatment decisions. The ability to comprehend context and semantics
within medical texts can aid clinicians in developing a reasonable differential diagnosis for the patient's symptoms. This aptitude allowed us to decode intricate medical jargon,
identify subtle nuances in symptoms, and establish correlations with relevant etiologies, even when symptom causation was not the primary focus of the source material. In doing so, on one
hand, we unearthed numerous etiologies that had not been previously documented in the reviews or the medical books, as potential causes for specific symptoms and on the other hand provided
good precision in finding etiologies compared to the traditional tools used today while creating a big list of differential diagnoses for a set of symptoms. Additionally, like most AI
models, our tool can substantially alleviate the cognitive burden on medical professionals by automating the initial phases of symptom analysis and etiology identification. This automation
empowers healthcare providers to redirect their expertise toward more critical tasks, such as patient interaction, treatment planning, and decision-making, ultimately bolstering the overall
efficiency and effectiveness of healthcare delivery. An important point at this time is that while AI-powered models undoubtedly offer numerous advantages in identifying symptom etiologies
and enhancing healthcare delivery, it is essential to recognize that a significant portion of the population may harbour reservations regarding the integration of AI into their healthcare.
According to a study conducted by the Pew Research Center, a noteworthy 60% of Americans express discomfort at the prospect of their healthcare provider relying on AI for their medical
care36. Our research endeavours have demonstrated that a collaboration between AI developers and medical professionals along with a synergistic approach, merging generative models like
ChatGPT with Syntactic patterns, yields both precise and extensive results. This amalgamation of AI and conventional methodologies could potentially serve as a crucial step in building trust
within the population when it comes to embracing AI models within the medical domain. As the healthcare sector continues to explore the potential of AI, it becomes increasingly vital to
leverage these tools effectively to not only enhance medical practice but also to foster confidence among patients and healthcare consumers. In our research, we highlighted the profound
relevance of AI in the realm of medical practice, particularly in the important task of identifying symptom etiologies. This cutting-edge technology presents a multitude of advantages when
compared to the traditional methods commonly employed by physicians today. DATA AVAILABILITY The datasets generated and/or analyzed during the current study are available from the
corresponding author on reasonable request. REFERENCES * Evidence-Based Medicine with UpToDate. https://www.wolterskluwer.com/en/solutions/uptodate/about/evidence-based-medicine. * Abulaish,
M., Parwez, M. A. & Jahiruddin, J. DiseaSE: A biomedical text analytics system for disease symptom extraction and characterization. _J. Biomed. Inf._ 100, 103324 (2019). Article Google
Scholar * Feng, Q., Gui, Y., Yang, Z., Wang, L. & Li, Y. Semisupervised learning based disease-symptom and symptom-therapeutic substance relation extraction from biomedical literature.
_Biomed. Res. Int._ 2016, 3594937 (2016). Article PubMed PubMed Central Google Scholar * Hassan, M., Makkaoui, O., Coulet, A. & Toussain, Y. Extracting disease-symptom relationships
by learning syntactic patterns from dependency graphs. in _Proc. BioNLP 15_ 184 (Association for Computational Linguistics, 2015). * Luo, X., Gandhi, P., Storey, S. & Huang, K. A deep
language model for symptom extraction from clinical text and its application to extract COVID-19 symptoms from social media. _IEEE J. Biomed. Health Inf._ 26, 1737–1748 (2022). Article
Google Scholar * Magge, A., Weissenbacher, D., Oâ Connor, K., Scotch, M. & Gonzalez-Hernandez, G. SEED: Symptom Extraction from English Social Media Posts using Deep Learning and
Transfer Learning. _medRxiv_ (2022). * Xia, E. _et al._ Mining disease-symptom relation from massive biomedical literature and its application in severe disease diagnosis. _AMIA Annu. Symp.
Proc._ 2018, 1118–1126 (2018). PubMed PubMed Central Google Scholar * Gupta, S. & Manning, C. Improved Pattern Learning for Bootstrapped Entity Extraction. in _Proc. Eighteenth Conf.
Comput. Nat. Lang. Learn._ 98–108 (Association for Computational Linguistics, 2014). * Vacareanu, R., Bell, D. & Surdeanu, M. PatternRank: Jointly ranking patterns and extractions for
relation extraction using graph-based algorithms. in _Proc. First Workshop Pattern-Based Approaches NLP Age Deep Learn._ 1–10 (International Conference on Computational Linguistics, 2022). *
Mallen, A., Asai, A., Zhong, V., Das, R., Khashabi, D. & Hajishirzi, H. When Not to Trust Language Models: Investigating Effectiveness of Parametric and Non-Parametric Memories. in
_Proc. 61st Annu. Meet. Assoc. Comput. Linguist. Vol. 1 Long Pap._ 9802–9822 (Association for Computational Linguistics, 2023). https://doi.org/10.18653/v1/2023.acl-long.546 * Kandpal, N.,
Deng, H., Roberts, A., Wallace, E. & Raffel, C. Large Language Models Struggle to Learn Long-Tail Knowledge. in _Proc. 40th Int. Conf. Mach. Learn._ (eds. Krause, A., Brunskill, E., Cho,
K., Engelhardt, B., Sabato, S. & Scarlett, J.) 202, 15696–15707 (PMLR, 2023). * Huang, C.-C. & Lu, Z. Community challenges in biomedical text mining over 10 years: Success, failure
and the future. _Brief. Bioinform._ 17, 132–144 (2016). Article PubMed Google Scholar * Zhao, S., Su, C., Lu, Z. & Wang, F. Recent advances in biomedical literature mining. _Brief.
Bioinform_. 22 (2021). * Symptom Checker with Body from WebMD—Check Your Medical Symptoms. _WebMD_. https://symptoms.webmd.com/ * Symptom Checker, Health Information and Medicines Guide.
https://patient.info/. * Symptom Checker|Isabel—The symptom checker doctors use. https://symptomchecker.isabelhealthcare.com. * Ramnarayan, P., Kulkarni, G., Tomlinson, A. & Britto, J.
ISABEL: A novel Internet-delivered clinical decision support system. _Curr. Perspect. Healthc. Comput._ 245–256 (2004). * Lagunes-García, G. _et al._ DISNET: A framework for extracting
phenotypic disease information from public sources. _PeerJ_ 8, e8580 (2020). Article PubMed PubMed Central Google Scholar * García del Valle, E. P. _et al._ Disease networks and their
contribution to disease understanding: A review of their evolution, techniques and data sources. _J. Biomed. Inf._ 94, 103206 (2019). Article Google Scholar * Zhou, X., Menche, J.,
Barabási, A.-L. & Sharma, A. Human symptoms–disease network. _Nat. Commun._ 5, 1–10 (2014). Article Google Scholar * Doğan, R. I., Leaman, R. & Lu, Z. NCBI disease corpus: A
resource for disease name recognition and concept normalization. _J. Biomed. Inf._ 47, 1–10 (2014). Article Google Scholar * Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C.-H.,
Leaman, R., Davis, A. P., Mattingly, C. J., Wiegers, T. C. & Lu, Z. BioCreative V CDR task corpus: A resource for chemical disease relation extraction. _Database_ 2016, (2016). *
Steinkamp, J. M., Bala, W., Sharma, A. & Kantrowitz, J. J. Task definition, annotated dataset, and supervised natural language processing models for symptom extraction from unstructured
clinical notes. _J. Biomed. Inf._ 102, 103354 (2020). Article Google Scholar * Causes of persistent and intractable hiccups. _UpToDate_.
https://www.wolterskluwer.com/en/solutions/uptodate/about/evidence-based-medicine. * Joseph, A. & Samant, H. in _StatPearls_ (StatPearls Publishing, 2023).
http://www.ncbi.nlm.nih.gov/books/NBK544252/. * Outpatient evaluation of the adult with chest pain—UpToDate.
https://www.uptodate.com/contents/outpatient-evaluation-of-the-adult-with-chest-pain?search=chest%20pain%20differential%20diagnosis&source=search_result&selectedTitle=2~150&usage_type=default&display_rank=2.
* Taub-Tabib, H., Shlain, M., Sadde, S., Lahav, D., Eyal, M., Cohen, Y. & Goldberg, Y. Interactive extractive search over biomedical corpora. (2020). * Clothiaux, D. & Starzl, R.
Extractive Search for Analysis of Biomedical Texts. in _Proc. 45th Int. ACM SIGIR Conf. Res. Dev. Inf. Retr._ 3386–3387 (Association for Computing Machinery, 2022). * Shlain, M., Taub-Tabib,
H., Sadde, S. & Goldberg, Y. Syntactic search by example. (2020). * Schwartz, A. S. & Hearst, M. A. in _Biocomput. 2003_ 451–462 (WORLD SCIENTIFIC, 2002). * Sung, M., Jeon, H., Lee,
J. & Kang, J. Biomedical entity representations with synonym marginalization. (2020). * Davis, A. P., Wiegers, T. C., Rosenstein, M. C. & Mattingly, C. J. MEDIC: A practical disease
vocabulary used at the Comparative Toxicogenomics Database. _Database_ 2012, bar065 (2012). Article PubMed PubMed Central Google Scholar * Ravfogel, S., Pyatkin, V., Cohen, A. D. N.,
Manevich, A., & others. Retrieving texts based on abstract descriptions. _ArXiv Prepr. ArXiv_ (2023). * Asai, A., Min, S., Zhong, Z. & Chen, D. Retrieval-based Language Models and
Applications. in _Proc. 61st Annu. Meet. Assoc. Comput. Linguist. Vol. 6 Tutor. Abstr._ 41–46 (Association for Computational Linguistics, 2023). * Dai, N., Xu, D., Zhong, X., Li, L., Ling,
Q. & Bu, Z. Build infrastructure in publishing scientific journals to benefit medical scientists. _Chin. J. Cancer Res. Chung-Kuo Yen Cheng Yen Chiu_ 26, 119–123 (2014). * How Americans
View Use of AI in Health Care and Medicine by Doctors and Other Providers|Pew Research Center.
https://www.pewresearch.org/science/2023/02/22/60-of-americans-would-be-uncomfortable-with-provider-relying-on-ai-in-their-own-health-care/. Download references ACKNOWLEDGEMENTS We thank
Layan Farraj, Heba Dahli, and Fatima Edrees for their participation in the annotation tasks, and their assistance and advice. Y.S. was funded by the Israeli Science Foundation Grant #901/19.
Y.S. received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme, Grant agreement No. 802774 (iEXTRACT). AUTHOR
INFORMATION AUTHORS AND AFFILIATIONS * Allen Institute for AI, Seattle, USA Hillel Taub-Tabib, Micah Shlain, Menny Pinhasov, Mark Polak, Aryeh Tiktinsky, Sigal Rahamimov, Dan Bareket, Ben
Eyal, Moriya Kassis & Yoav Goldberg * Faculty of Biomedical Engineering, Technion, Haifa, Israel Yosi Shamay * Computer Science Department, Bar Ilan University, Ramat Gan, Israel Yoav
Goldberg * Technion Faculty of Medicine Library and Rambam Health Campus Library, Haifa, Israel Tal Kaminski Rosenberg * Institute for Pain Medicine, Rambam Health Campus, Haifa, Israel
Simon Vulfsons & Maayan Ben Sasson * Alan Edwards Pain Management Unit, McGill University Health Centre, Montreal, QC, Canada Maayan Ben Sasson Authors * Hillel Taub-Tabib View author
publications You can also search for this author inPubMed Google Scholar * Yosi Shamay View author publications You can also search for this author inPubMed Google Scholar * Micah Shlain
View author publications You can also search for this author inPubMed Google Scholar * Menny Pinhasov View author publications You can also search for this author inPubMed Google Scholar *
Mark Polak View author publications You can also search for this author inPubMed Google Scholar * Aryeh Tiktinsky View author publications You can also search for this author inPubMed Google
Scholar * Sigal Rahamimov View author publications You can also search for this author inPubMed Google Scholar * Dan Bareket View author publications You can also search for this author
inPubMed Google Scholar * Ben Eyal View author publications You can also search for this author inPubMed Google Scholar * Moriya Kassis View author publications You can also search for this
author inPubMed Google Scholar * Yoav Goldberg View author publications You can also search for this author inPubMed Google Scholar * Tal Kaminski Rosenberg View author publications You can
also search for this author inPubMed Google Scholar * Simon Vulfsons View author publications You can also search for this author inPubMed Google Scholar * Maayan Ben Sasson View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Conceptualization: S.V, M.B, H.T Methodology: Y.S, T.K, M.B, S.V, H.T Data curation: H.T, D.B, B.E,
Y.G, M.S, M.Pi, M.Po, A.T, S.R Formal analysis: H.T, M.B Writing – Original Draft Preparation M.B, H.T Writing – Review & Editing: S.V, Y.S, T.K Visualization: M.K Supervision: M.S, S.V,
Y.G Project Administration: M.B, H.T, CORRESPONDING AUTHOR Correspondence to Maayan Ben Sasson. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests.
ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RIGHTS AND PERMISSIONS
OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or
format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or
other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not
included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission
directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Taub-Tabib, H., Shamay, Y., Shlain, M. _et al._ Identifying symptom etiologies using syntactic patterns and large language models. _Sci Rep_ 14, 16190 (2024).
https://doi.org/10.1038/s41598-024-65645-6 Download citation * Received: 19 December 2023 * Accepted: 21 June 2024 * Published: 13 July 2024 * DOI: https://doi.org/10.1038/s41598-024-65645-6
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy
to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
