Effects of low-intensity pulsed ultrasound on the microorganisms of expressed prostatic secretion in patients with iiib prostatitis
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT To detect and analyze the changes of microorganisms in expressed prostatic secretion (EPS) of patients with IIIB prostatitis before and after low-intensity pulsed ultrasound (LIPUS)
treatment, and to explore the mechanism of LIPUS in the treatment of chronic prostatitis (CP). 25 patients (study power was estimated using a Dirichlet-multinomial approach and reached
96.5% at α = 0.05 using a sample size of 25) with IIIB prostatitis who were effective in LIPUS treatment were divided into two groups before and after LIPUS treatment. High throughput
second-generation sequencing technique was used to detect and analyze the relative abundance of bacterial 16 s ribosomal variable regions in EPS before and after treatment. The data were
analyzed by bioinformatics software and database, and differences with P < 0.05 were considered statistically significant. Beta diversity analysis showed that there was a significant
difference between groups (P = 0.046). LEfSe detected four kinds of characteristic microorganisms in the EPS of patients with IIIB prostatitis before and after LIPUS treatment. After
multiple comparisons among groups by DESeq2 method, six different microorganisms were found. LIPUS may improve patients’ clinical symptoms by changing the flora structure of EPS, stabilizing
and affecting resident bacteria or opportunistic pathogens. SIMILAR CONTENT BEING VIEWED BY OTHERS URINARY MICROBIOTA AND PROSTATIC DISEASES: THE KEY FOR THE LOCK? A SYSTEMATIC REVIEW
Article 08 October 2022 OVERVIEW OF SEMINAL FLUID BIOMARKERS FOR THE EVALUATION OF CHRONIC PROSTATITIS: A SCOPING REVIEW Article 29 November 2021 IDENTIFICATION OF AN INTESTINAL MICROBIOTA
ENTEROTYPES IN AGEING MAN DIAGNOSED WITH BENIGN PROSTATIC HYPERPLASIA (BPH) Article Open access 04 May 2025 INTRODUCTION Chronic prostatitis (CP) usually refers to a prostate disease caused
by various complex causes, with urethral irritation and chronic pelvic pain as the primary clinical manifestations. It is among the most common chronic diseases in young and middle-aged
men1. Because it is difficult to treat and easy to relapse, it can severely affect the quality of life of patients and their mental status2,3. Unfortunately, the incidence of CP has been
increasing in recent years. It has been reported that the incidence of CP in China has reached 6.0–32.9%4. Unfortunately, the pathogenesis of CP is still not very clear. Some scholars have
recently found that CP may be closely related to some potential pathogenic microorganisms5,6,7. Currently, the clinical treatment of CP is still dominated by therapeutic medicines.
Medication treatment can alleviate the clinical symptoms of patients to some extent. However, for some patients, especially IIIB prostatitis, the effect of medication treatment is poor, and
the recurrence rate is high8. Consequently, the side effects of medication treatment are difficult to avoid and may sometimes even outweigh the potential therapeutic effect on the disease9.
Therefore, as a new treatment for CP, clinicians, and patients are now considering low-intensity pulsed ultrasound (LIPUS). In recent years, many reports have been published on the efficacy
and safety of LIPUS in treating IIIB prostatitis10,11,12. With the increasingly extensive application of LIPUS, people are beginning to pay more attention to its effective mechanism, which
is of great significance not only for further promoting the application of LIPUS but also for understanding its possible side effects and its efficacy. Because of the added research on the
relationship between pathogenic microorganisms and CP in recent years, we aimed to explore the possible mechanism of LIPUS in the treatment of IIIB prostatitis from the point of view of the
effect of LIPUS on microorganisms in patients’ expressed prostatic secretion (EPS). MATERIALS AND METHODS STUDY POPULATION Prior to study initiation, study power was estimated using a
Dirichlet-multinomial approach and reached 96.5% at α = 0.05 using a sample size of 2513. In strict accordance with the classification criteria of prostatitis established by the National
Institutes of Health (NIH), we selected 25 patients with IIIB prostatitis who were only effectively treated with LIPUS (the total score of NIH-chronic prostatitis symptom index (CPSI)
decreased by four or more points) in Xiangya Third Hospital of Central South University from January 2022 to June 2022. All subjects were between the ages of 20 and 40 (CP/CPPS tends to
occur more frequently in middle-aged and young adult males). This study was approved by the Ethics Committee of Xiangya Third Hospital of Central South University (Grant No. 22069), and the
subjects' informed consent was obtained. SELECTION CRITERIA The inclusion criteria were as follows: (i) age was between 20 and 40 years old; (ii) NIH was classified as CPPS (IIIB), the
clinical symptoms lasted more than 3 months, and the total score of CPSI was more than 10 points9; (iii) only received LIPUS treatment and regularly followed two courses, and the total CPSI
score decreased to or more than four points; (iv) did not take any antibiotics within 3 months before LIPUS treatment; (v) the patient signed informed consent. The exclusion criteria were
designated as follows: (i) urinary tract infection; (ii) history of urinary cancer, surgery, radiotherapy, systemic chemotherapy; (iii) unilateral testicular pain, active urethral stricture
or bladder stone with pelvic symptoms, or any other urinary disease associated with lower urinary tract symptoms, any neurological disease or disorder affecting the bladder; (iv) did not
receive regular LIPUS treatment for two courses, or while receiving LIPUS treatment, took other therapeutic medicines or took antibiotics within 3 months before LIPUS treatment; (v) did not
sign informed consent. GROUPING AND SPECIMEN COLLECTION We analyzed symptoms of our patients before and after LIPUS treatment. During the treatment of LIPUS, the CP treatment mode was
selected, and the patients were instructed to take the bladder lithotomy position and fully expose the perineum and pubic symphysis. The treatment head A and B of the therapeutic instrument
were fixed in the perineum and pubic symphysis, respectively. The energy intensity was set at 1.25 w/cm2. Each treatment lasted for 10 min, once every other day, and five consecutive
treatments were taken as a course of treatment. EPS samples of 25 subjects were collected before and after two courses of treatment in 30 min. Before collecting, we asked the patient to
empty their bladder. We then cleaned the urethral orifice with sterile normal saline and wiped the residual liquid near the urethral orifice with aseptic gauze. Aseptically collected EPS
samples were obtained by the same urologist from all patients included in the study. All specimens were stored in the refrigerator at − 80 ℃ immediately after collection. INSTRUMENTS AND
REAGENTS The main instrument LIPUS therapeutic instrument model is LY-ED01, purchased from Hunan Lanyue Medical Technology Co Ltd. (Changsha, China); Nucleic acid electrophoresis instrument
model DYCP-32C, agarose level electrophoresis instrument, purchased from Liuyi instrument Factory (Beijing, China); Covaris ultrasonic crusher Covaris S2 System, purchased from Massachusetts
(USA); Qubit Fluorometric Quantification Qubit 2.0, purchased from Life Technologies (CA, USA); Bioanalyzer Agilent 2100, purchased from Agilent Technologies Co Ltd. (USA); The PCR
instrument is T100PCR, purchased from Bio-Rad (USA); The sequencer is NovaSeq 6000, purchased from Illumina, San Diego (CA, USA), and the product purification kit is Qiagen gel recovery kit
purchased from Qiagen Company (DUS, GER). LIBRARY CONSTRUCTION Total genomic DNA was extracted from EPS samples using the CTAB method. PCR amplified the V3+V4 variable region of 16 s
ribosomal RNA (rRNA) gene with the primers 341F (5ʹ-CCTAYGGGRBGCASCAG-3ʹ) and 806R (5ʹ-GGACTACNNGGGTATCTAAT-3ʹ), respectively. The samples were mixed with the same concentration according to
the concentration of PCR products. Then the PCR products were purified by agarose gel electrophoresis with 1 × TAE concentration of 2%, and the target bands were recovered by tapping. We
used the Illumina TruSeq® DNA PCR-Free sample preparation kit library building kit (Illumina, USA) to build the library and add the index code. The constructed library was evaluated by Qubit
quantification and library detection. Finally, the NovaSeq 6000 PE250 was used for sequencing, and the peer reads of 250 bp were obtained. BIOINFORMATICS ANALYSIS The analysis process was
mainly done with reference to the “Atacama soil microbiome tutorial” tutorial in the Qiime2 documentation (https://docs.qiime2.org/2019.1/). The original sequence fastq file was imported by
qiime tools import plug-in and converted into a file format that can be processed later by QIIME2. The QIIME2 dada2 plug-in was used to complete the quality control steps, pruning,
denoising, splicing, and removing chimera, and finally, we obtained the feature sequence table14. Then the QIIME2 feature-classifier plug-in was used to compare the representative sequences
of ASV to the pre-trained GREENGENES database with 99% similarity of version 13_8 (pruning the database to the area of V3V4 according to 341F/806R primer pairs), to get the species
classification information table15. The QIIME2 feature-table plug-in removed all contaminated mitochondrial and chloroplast sequences. By using the “mixOmics” software package of R (v3.1.1)
and partial least squares discriminant analysis (PLS-DA) multivariate statistical analysis method, according to the values of several variables observed or measured, how to classify the
research objects and reveal the relationship between microbial communities and sample categories16. The diversity matrix is calculated by using the QIIME2 core-diversity plug-in. The Alpha
diversity index evaluates the diversity degree of the sample itself at the feature sequence level. Notably, the Alpha diversity index includes observed operational taxonomic units (OUTs),
Chao, Faith’s phylogenetic diversity, Shannon, and Simpson. The Beta diversity index evaluated the difference in microbial community structure among samples. Beta diversity index includes
Bray Curtis and unweighted UniFrac and weighted UniFrac index, and the results are shown by PCoA and NMDS maps17. After obtaining the overall Beta diversity index, we combined the grouping
information. The PERMANOVA and ANOSIM methods were used to compare whether there were significant differences in microbial composition and structure among different sample groups.
Microorganisms with characteristics and different abundance among groups were identified by LEfSe and DEseq2 methods18,19. LEfSe method is based on the relative abundance table, a
combination of nonparametric test and linear discriminant analysis and is suitable for the flora abundance difference test. It can find the characteristic microbes of each group (LDA > 2
& LDA > 4), that is, the microbes with high abundance in this group compared to other groups. The DESeq2 method can be used to find microbes with significant differences between
groups by making multiple comparisons between groups. Unless otherwise noted, the parameters used in the above analysis are default settings. ETHICS APPROVAL AND CONSENT TO PARTICIPATE The
study was conducted in accordance with the Declaration of Helsinki, and approved by Institutional Review Board (IRB) of The Third Xiangya Hospital of Central South University (Grant No.
22069). Along with the subjects’ informed consent was obtained. RESULTS PATIENTS’ INFORMATION A total of 25 patients with IIIB prostatitis treated with LIPUS were enrolled in this study. The
total score of CPSI was more than 10 points before treatment and decreased by four or more points after treatment (Table 1). OTU ABUNDANCE ANALYSIS A total of 7,385,355 readable fragments
were obtained from the valid data of 50 samples. After merging tag clustering, all valid sequences are clustered/denoised, and 17,535 OTUs were obtained according to 16 s rRNA data. We can
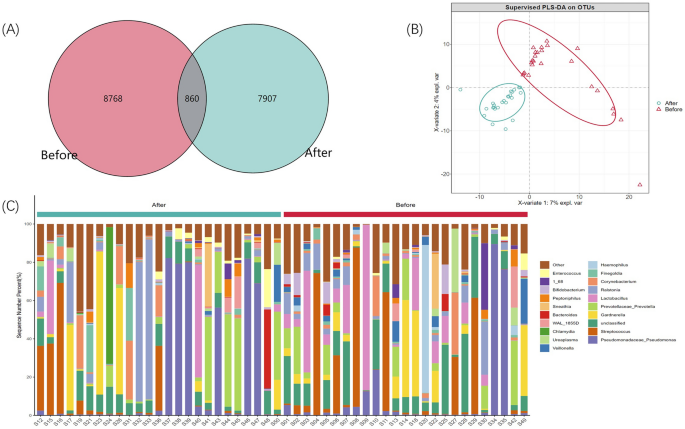
find the unique or common OTUs by comparing the OTUs between samples. By drawing a Venn diagram to analyze the unique or common OTU between different sample groups, which directly shows the
composition similarity and overlap of samples between groups at the OTU level (Fig. 1A). At the same time, we carried out a PLS-DA analysis, using the R language mixOmics package, of all OTU
with an abundance greater than 10 and provided coordinate diagrams (Fig. 1B). From the Wayne diagram and PLS-DA analysis coordinate chart, it can be seen that there are differences in the
structure of microflora in the EPS of patients with IIIB prostatitis before and after LIPUS treatment. ANALYSIS OF TAXONOMIC ANNOTATIONS The species annotation information was obtained by
selecting the representative sequence of OTU and comparing it with the GREENGENES database (GREENGENES Database 13_8 version)20. Based on the species annotation information, the annotations
were removed as chloroplasts, mitochondria, and OTU that cannot be annotated to the boundary level and the sequences they contain. Based on OUT’s absolute abundance and annotated
information, 39 phylum, 100 classes, 162 orders, 236 families, 408 genera, and 245 species were annotated in this study. Figure 1C shows the distribution of each group’s top 20 species of
relative abundance at the genus level. In contrast, Figs. S1, S2, S3, S4, S5 shows the relative distribution of the species in the top 20 of relative abundance in each group at the phylum,
class, order, family, and species level, respectively. HEATMAP ANALYSIS To study the similarity between different samples, the species concerned about the classification level (the first 20
of the default absolute abundance of species) are selected to realize sample clustering to investigate the similarities or differences between different samples or groups. We then carried on
the horizontal clustering from the two aspects of classification information and the difference between samples to find the aggregation law of species or samples. In this study, heatmap
cluster analysis was carried out at the phylum, class, order, family, genus (Fig. 2A), and species level. According to the 16 s rRNA sequence, the top five bacteria were _Pseudomonas_,
_Prevotella_, _Streptococcus_, _Ralstonia_, and _Lactobacillus_. The phylogenetic analysis of the representative sequence of OTU was carried out by using the R language ggtree package (each
genus selects an OTU with the highest abundance as the representative OTU, and then the top 50 genera with the highest abundance), the evolutionary tree was drawn, and the heat map was
visualized with the absolute abundance of OTU in each group (Fig. 2B). SAMPLE-BASED RAREFACTION ANALYSIS Alpha diversity analysis was used to evaluate the microbial diversity of the sample
itself, which was completed by the qiime2 diversity plug-in. The index of a single sample of sparse curves with different α diversity shows that the sequencing depth covers all species in
the sample (Fig. 3F). Microbial diversity was described by observed features, Chao, Faith’s phylogenetic diversity, Shannon, and Simpson. After obtaining the overall Alpha diversity index,
combined with grouping information, Wilcox test was used to compare the significant differences between groups accurately. The results showed no significant difference in species diversity
between groups, as shown in Table 2. Figure 3A–E is a box chart of Alpha diversity between groups. Beta diversity analysis was used to evaluate samples’ differences in microbial community
structure. Bray Curtis obtained the whole Beta diversity index, and the microbial community structure between groups was compared by the PERMANOVA method combined with grouping information.
The results demonstrated a significant difference (P = 0.046), as shown in Table 3. Furthermore, Fig. 4A–C shows the groups’ Beta diversity PCoA and NMDS diagrams. LEFSE ANALYSIS LEfSe
analysis showed that when LDA Score was equal to two at the genus level, 34 kinds of characteristic microorganisms were found in the EPS of patients before and after LIPUS treatment,
including 29 before treatment and five after treatment (Fig. 5B). When the LDA score was equal to four at the genus level, four characteristic microorganisms were found in the EPS of
patients with LIPUS before-treatment group, namely, _Lactobacillales_, _Bacilli_, _Veillonella_, and _Firmicutes_ (Fig. S6). No characteristic microorganisms were found in the
after-treatment group. SIGNIFICANCE ANALYSIS OF INTERGROUP DIFFERENCES By using the R language DESeq2 package, it was found that there were significant differences in six kinds of bacteria
at genus level between patients before and after LIPUS treatment groups (log2FC > 2 & P < 0. 05). There were significant differences in _Veillonella_, _Methyloversatilis_,
_Morganella_, _Bifidobacterium_ and _Ureaplasma_ in EPS before LIPUS treatment. There was a significant difference in _porphyromonas_ in EPS after LIPUS treatment (Fig. 5A). DISCUSSION
Prostatitis is the third most common disease in the genitourinary system. NIH classifies prostatitis into four types, of which type III chronic abacterial prostatitis/chronic pelvic pain
syndrome (CP/CPPS) is the most common type, accounting for 80–90% of prostatitis21. According to EPS, semen, and urine samples after a prostate massage, CP/CPPS can be divided into
inflammatory (IIIA) and non-inflammatory types (IIIB)22. Although no pathogens were found in CP/CPPS patients undergoing routine EPS bacterial culture, many researchers still consider
infection as an important pathogenic factor. Mycoplasma, chlamydia, trichomonas, candida, viruses and parasites, and other pathogenic microorganism infections have been reported to be
associated with the pathogenesis of CP/CPPS23,24,25. At present, the primary treatment of CP/CPPS is medication therapy. Commonly used medicines in the clinic include α-receptor blockers,
plant preparations, non-steroidal anti-inflammatory analgesics, and muscarinic receptor blockers. However, patients often report that the medication treatment is ineffective, the recurrence
rate is high, and they are eager to receive more effective treatment11. It is under this background that LIPUS was born. The efficacy and safety of LIPUS in treating CP/CPPS, especially in
the treatment of IIIB prostatitis, has been confirmed by many clinical researchers inland and abroad1,11,12,26,27. With the popularization and application of LIPUS, the effective mechanism
of CP treatment has been paid more attention to by patients and doctors. This study investigated 25 patients with IIIB prostatitis whose symptoms were significantly improved after LIPUS
treatment. The microorganisms in their EPS before and after treatment were detected and analyzed. The results showed that _Pseudomonas, Prevotella_, _Streptococcus_, _Ralstonia,_ and
_Lactobacillus_ were the most abundant bacteria in EPS. The structure and abundance of microflora in EPS of patients with IIIB prostatitis changed significantly before and after LIPUS
treatment. The abundance of _Veillonella_, _Methyloversatilis_, _Morganella_, _Bifidobacterium_, _Ureaplasma_, _Lactobacillales_, _Bacilli_, and _Firmicutes_ in EPS decreased significantly
after LIPUS treatment (Fig. 5A and Fig. S6). It is important to emphasize that while our study results show the presence of multiple bacterial species in EPS from IIIB prostatitis patients,
this is due to the use of advanced high-throughput sequencing technology. These bacteria would not have been detected through routine bacterial culture processes of EPS from IIIB prostatitis
patients. As antimicrobial therapy was not clinically indicated, we solely utilized LIPUS as adjunctive therapy. With the continuous development of detection technology, researchers have
found microbial flora in many parts and organs of the human body. Notably, microbes have been detected in men’s urethra, prostate, and testes28,29,30. Microorganisms in human organs usually
have unique flora structures and microecology29. As a colorless milky fluid continuously secreted by prostate epithelial cells, EPS can reflect the function and state of the prostate to some
extent31. In support of this, Davidsson et al. found propionibacterium acne mainly in prostate cancer tissues and normal tissues32. Feng et al. found that prostate cancer tissues and normal
tissues are rich in Escherichia coli, Propionibacterium acne, Acinetobacter, and Pseudomonas at the macro-genomic level33. Lee et al. isolated _Enterococcus faecalis_ from EPS samples of CP
patients34. Fang et al. used high-throughput sequencing technology to detect EPS samples from CP patients and normal healthy men and found a significant difference in the composition of the
two flora as a whole29. Furthermore, we also detected a variety of bacteria in the EPS of patients, confirming the prevalence of microbial phenomena in EPS. Interestingly, we found that the
microflora in the EPS of the patients with IIIB prostatitis in the study changed significantly before and after LIPUS treatment groups and that the species and abundance of microorganisms
decreased significantly after treatment. The reason for the analysis may be that some of the bacteria in EPS were destroyed by ultrasound produced by LIPUS. As early as 1927, Wood and others
confirmed that ultrasound could have a fatal effect on microorganisms35. Follow-up studies also found that ultrasound can kill microorganisms, whether used alone or in combination with
other means, whether high intensity or low intensity36,37,38. However, the patients with IIIB prostatitis in this study only received LIPUS treatment, and the symptoms of CP were
significantly improved. Therefore, we hypothesize that during the treatment of IIIB prostatitis, some microorganisms in the EPS of patients with IIIB prostatitis are destroyed by the
ultrasound emitted by LIPUS, resulting in the change of flora structure. Notably, this may be one of the reasons for the obvious improvement of CP symptoms after LIPUS treatment. In this
study, the EPS of patients was collected through urethral excretion. The microorganisms in EPS reflect the microorganisms in the prostate tissue and include some urethral colonization
microorganisms. Importantly, we study EPS as a whole. Our research utilized advanced detection methods and scientific, statistical analyses methods to demonstrate that the structure and
abundance of flora in EPS are changed following LIPUS treatment. However, for patients with CP/CPPS, it is not very clear whether there are specific pathogens in EPS and whether the changes
in the structure and abundance of resident microflora or opportunistic pathogens in EPS are directly related to the clinical symptoms of patients. In theory, when the microecology of
microflora in EPS changes under the action of some influencing factors, it may lead to the imbalance of normal flora or opportunistic pathogens, resulting in the occurrence and development
of inflammation7. Following LIPUS treatment, some resident bacteria or opportunistic pathogens may be inhibited or eliminated to a certain extent, so the structure and abundance of bacteria
in EPS tend to be balanced and stable. Thus, reducing the clinical symptoms of patients to a certain extent. However, the profound relationship between the structure and abundance of
microflora in EPS and inflammatory factors and the pathogenesis of CP remains to be further studied. Moreover, normal microbial communities within individuals may vary due to differences in
race, diet, culture, and other factors across regions. How the flora distribution of patients in different regions will change before and after LIPUS treatment needs to be further confirmed
by multicenter collaborative research. CONCLUSION In patients with IIIB prostatitis whose symptoms were significantly improved after LIPUS treatment, there were significant changes in the
structure of flora in EPS before and after treatment. LIPUS may improve the clinical signs of patients with IIIB prostatitis by changing the flora structure of EPS, stabilizing and affecting
resident bacteria or opportunistic pathogens. DATA AVAILABILITY The data presented in the study are deposited in the NCBI repository, accession number PRJNA1050891. REFERENCES * Wang, Y.
B., He, D. H. & Yang, H. T. Clinical observation of low energy extracorporeal shock wave in the treatment of type IIIB prostatitis. _Chin. Community Phys._ 34(22), 77–79 (2018). ADS
Google Scholar * Song, W. J., Liu, X. H. & He, L. Y. Research progress on the relationship between prostate disease and erectile dysfunction. _J. Urol._ 11(04), 35–38 (2019). Google
Scholar * Banyra, O., Ivanenko, O., Nikitin, O. & Shulyak, A. Mental status in patients with chronic bacterial prostatitis. _Cent. Eur. J. Urol._ 66(1), 93–100 (2013). Article Google
Scholar * Yu, X. J. & Gao, Q. H. Guidelines for multidisciplinary diagnosis and treatment of chronic prostatitis with integrated traditional Chinese and western medicine. _Chin. J.
Androl._ 26(04), 82–89 (2020). CAS Google Scholar * Mo, X. _et al._ Prevalence and correlates of _Mycoplasma genitalium_ infection among prostatitis patients in Shanghai, China. _Sex.
Health_ 13, 474–479 (2016). Article Google Scholar * Xiao, J. Q. _et al._ Study on pathogenic microorganisms of prostatic fluid in patients with refractory chronic prostatitis. _Chin. J.
Androl._ 10, 5 (2010). Google Scholar * Wu, Y. _et al._ Screening for chronic prostatitis pathogens using high-throughput next-generation sequencing. _Prostate_ 80, 577–587 (2020). Article
CAS PubMed PubMed Central Google Scholar * Franco, J. V. _et al._ Non-pharmacological interventions for treating chronic prostatitis/chronic pelvic pain syndrome. _Cochrane Database
Syst. Rev._ 1, CD012551 (2018). PubMed Google Scholar * Franco, J. V. _et al._ Pharmacological interventions for treating chronic prostatitis/chronic pelvic pain syndrome. _Cochrane
Database Syst. Rev._ 10, CD012552 (2019). PubMed Google Scholar * Yuan, P. _et al._ Efficacy of low-intensity extracorporeal shock wave therapy for the treatment of chronic
prostatitis/chronic pelvic pain syndrome: A systematic review and meta-analysis. _Neurourol. Urodyn._ 38, 1457–1466 (2019). Article PubMed Google Scholar * Zhu, X. D. _et al._ Clinical
analysis of low energy shock wave in the treatment of intractable type III B prostatitis. _J. Clin. Urol._ 10, 4 (2020). Google Scholar * Mykoniatis, I. _et al._ Evaluation of a
low-intensity shockwave therapy for chronic prostatitis type IIIb/chronic pelvic pain syndrome: A double-blind randomized sham-controlled clinical trial. _Prostate Cancer Prostatic Dis._ 24,
370–379 (2021). Article PubMed Google Scholar * La, R. P. S. _et al._ Hypothesis testing and power calculations for taxonomic-based human microbiome data. _PLoS One_ 7(12), e52078
(2012). ADS Google Scholar * Callahan, B. J. _et al._ DADA2: High-resolution sample inference from Illumina amplicon data. _Nat. Methods_. 13(7), 581–583.
https://doi.org/10.1038/nmeth.3869 (2016). PMID: 27214047; PMCID: PMC4927377. * Bokulich, N. A. _et al._ Optimizing taxonomic classification of marker gene amplicon sequences. _Microbiome_
https://doi.org/10.1186/s40168-018-0470-z (2018). Article PubMed PubMed Central Google Scholar * Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and
dispersion for RNA-seq data with DESeq2. _Genome Biol._ 15(12), 550 (2014). Article PubMed PubMed Central Google Scholar * Vázquez-Baeza, Y. _et al._ EMPeror: A tool for visualizing
high-throughput microbial community data. _Gigascience_ 2, 2047 (2019). Google Scholar * Rohart, F. _et al._ mixOmics: An R package for ‘omics feature selection and multiple data
integration. _PLoS Comput. Biol._ 13(11), e1005752 (2017). Article PubMed PubMed Central Google Scholar * Mandal, S. _et al._ Analysis of composition of microbiomes: A novel method for
studying microbial composition. _Microb. Ecol. Health Dis._ 26, 27663 (2015). PubMed Google Scholar * DeSantis, T. Z. _et al._ Greengenes, a chimera-checked 16S rRNA gene database and
workbench compatible with ARB. _Appl. Environ. Microbiol._ 72, 5069–5072 (2006). Article ADS CAS PubMed PubMed Central Google Scholar * Videcnik, Z. J. _et al._ Diagnosis and treatment
of bacterial prostatitis. _Acta Dermatovenerol. Alp Pannonica Adriat._ 24(2), 25–29 (2015). Google Scholar * Nickel, J. C. Prostatitis. _Can. Urol. Assoc. J._ 5(5), 306–315 (2011). Article
PubMed PubMed Central Google Scholar * Arora, H. C., Eng, C. & Shoskes, D. A. Gut microbiome and chronic prostatitis/chronic pelvic pain syndrome. _Ann. Transl. Med._ 5(2), 30
(2017). Article PubMed PubMed Central Google Scholar * Skerk, V. _et al._ Chronic prostatitis caused by _Trichomonas vaginalis_—Diagnosis and treatment. _J. Chemother._ 14(5), 537–538
(2002). Article CAS PubMed Google Scholar * Irajian, G. _et al._ Molecular detectionof _Ureaplasma urealyticum_ from prostate tissues using PCR-RFLP, Tehran, Iran. _Iran. J. Pathol._
11(2), 138–143 (2016). PubMed PubMed Central Google Scholar * Zimmermann, R., Cumpanas, A., Miclea, F. & Janetschek, G. Extracorporeal shock wave therapy for the treatment of chronic
pelvic pain syndrome in males: A randomised, double-blind, placebocontrolled study. _Eur. Urol._ 56, 418–424 (2009). Article PubMed Google Scholar * Marszalek, M., Berger, I. &
Madersbacher, S. Low-energy extracorporeal shock wave therapy for chronic pelvic pain syndrome: Finally, the magic bullet?. _Eur. Urol._ 56, 425–426 (2009). Article PubMed Google Scholar
* Cavarretta, I. _et al._ The microbiome of the prostate tumor microenvironment. _Eur Urol_ 72(4), 625–631 (2017). Article CAS PubMed Google Scholar * Fang, D. B. _Study on the Diversity
of Microflora in Semen and Prostate Massage Fluid of Patients With Type III Prostatitis_ (Zhejiang University, 2014). Google Scholar * Alfano, M. _et al._ Testicular microbiomein
azoospermic men-first evidence of the impact of an altered microenvironment. _Hum. Reprod._ 33(7), 1212–1217 (2018). Article CAS PubMed PubMed Central Google Scholar * Li, Y., Zhou, Y.
C. & Shang, X. J. Effects of reproductive system microflora on male health and related diseases. _J. Clin. Urol._ 27(11), 1030–1034 (2021). * Davidsson, S. _et al._ Frequency and typing
of Propionibacterium acnes in prostate tissue obtained from men with and without prostate cancer. _Infect. Agents Cancer_ 11, 26 (2016). Article Google Scholar * Feng, Y. _et al._
Metagenomic and metatranscriptomic analysis of human prostate microbiota from patients with prostate cancer. _BMC Genom._ 20(1), 146 (2019). Article Google Scholar * Lee, G. Chronic
prostatitis: A possible cause of hematospermia. _World J Mens Health_ 33(2), 103–108 (2015). Article PubMed PubMed Central Google Scholar * Wood, R. W. & Loomis, A. L. The physical
and biological effects of high-frequency sound-waves of great intensity. _Lond. Edinb. Dublin Philos. Mag. J. Sci._ 4, 417–436 (1927). Article CAS Google Scholar * Zhang, Y. I. _et al._
Hematoporphyrin monomethyl ether mediated sonodynamic antimicrobial chemotherapy on porphyromonas gingivalis in vitro. _Microb. Pathog._ 144, 104192 (2020). Article CAS PubMed Google
Scholar * Bhavya, M. L. & Hebbar, H. U. Sono-photodynamic inactivation of _Escherichia coli_ and _Staphylococcus aureus_ in orange juice. _Ultrason. Sonochem._ 57, 108–115 (2019).
Article CAS PubMed Google Scholar * Fan, L. _et al._ Sonodynamic antimicrobial chemotherapy: An emerging alternative strategy for microbial inactivation. _Ultrason. Sonochem._ 75(3),
105591 (2021). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS All work was completed at the department of urology, Central South University, The
Third Xiangya Hospital. The authors thank AiMi Academic Services (www.aimieditor.com) for the English language editing and review services. FUNDING LIPUS device and the two treatment heads
were provided by Hunan Lanyue medical Technology Co Ltd., Changsha, China, and this study was supported by the National Natural Science Foundation of China (No. 82101694). AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Department of Urology, The Third Xiangya Hospital, Central South University, No. 138 Tongzipo Road, Changsha City, 410013, Hunan Province, China Wei-Jie Song,
Ji-Wei Huang, Yuan Liu, Jie Wang, Wei Ding, Bin-Long Chen, Dong-Yi Peng, Zhi Long & Le-Ye He * Sexual Health Research Center, The Third Xiangya Hospital, Central South University,
Changsha, Hunan, China Wei-Jie Song, Ji-Wei Huang, Yuan Liu, Jie Wang, Wei Ding, Bin-Long Chen, Dong-Yi Peng, Zhi Long & Le-Ye He Authors * Wei-Jie Song View author publications You can
also search for this author inPubMed Google Scholar * Ji-Wei Huang View author publications You can also search for this author inPubMed Google Scholar * Yuan Liu View author publications
You can also search for this author inPubMed Google Scholar * Jie Wang View author publications You can also search for this author inPubMed Google Scholar * Wei Ding View author
publications You can also search for this author inPubMed Google Scholar * Bin-Long Chen View author publications You can also search for this author inPubMed Google Scholar * Dong-Yi Peng
View author publications You can also search for this author inPubMed Google Scholar * Zhi Long View author publications You can also search for this author inPubMed Google Scholar * Le-Ye
He View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS WS and JH initiated the research question and supervised all aspects of the study; YL
and JW contributed to data acquisition; WD and BC supervised the statistical procedures of the manuscript; WS wrote the first version of the manuscript; DP, ZL and LH reviewed this article.
All authors have read and agreed to the published version of the manuscript. CORRESPONDING AUTHOR Correspondence to Le-Ye He. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits
use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the
Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless
indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory
regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Song, WJ., Huang, JW., Liu, Y. _et al._ Effects of low-intensity pulsed ultrasound
on the microorganisms of expressed prostatic secretion in patients with IIIB prostatitis. _Sci Rep_ 14, 15368 (2024). https://doi.org/10.1038/s41598-024-66329-x Download citation * Received:
06 December 2023 * Accepted: 01 July 2024 * Published: 04 July 2024 * DOI: https://doi.org/10.1038/s41598-024-66329-x SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative KEYWORDS * Low-intensity pulsed ultrasound * IIIB prostatitis * High throughput * Second-generation sequencing * Expressed prostatic secretion * Microorganism
