Parp inhibitors elicit a cellular senescence mediated inflammatory response in homologous recombination proficient cancer cells
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Poly (ADP-ribose) polymerase (PARP) inhibitors have improved the prognosis of homologous recombination deficient (HRD) ovarian cancer (OC), while effective therapeutic strategies
for HR-proficient (HRP) OC still need to be established. This study investigates senescence-mediated inflammation as a novel mechanism of action for PARP inhibitors in HRP cancers.
Transcriptome analyses were performed in olaparib-treated HeLa cells as a HRP model. Interferon regulatory factor-Lucia luciferase (IRF-Luc) reporter activity was assessed. The effects of
PARP inhibitors on senescence-like phenotypes were assessed in seven HRP cancer cell lines, based on morphological changes, senescence-associated β-galactosidase (SA-β-GAL) activity,
cellular granularity, and senescence-associated secretory phenotype (SASP)-related gene expression. Peripheral blood mononuclear cell (PBMC) migration assays were also performed with the
conditioned medium in treatment with the PARP inhibitor. Transcriptome analyses revealed numbers of inflammatory cytokine- and chemokine-related pathways were significantly upregulated in
olaparib-treated HeLa cells, which were confirmed by IRF-Luc reporter assays. The PARP inhibitors induced senescent phenotypes in HRP cancer cell lines: flattened and enlarged morphology,
increased SA-β-GAL activity, elevated cellular granularity, and upregulated expressions of SASP-related genes (e.g., _IL1B_, _IL6_, and _CXCL10_). Furthermore, in vitro migration assays
revealed that PARP inhibitor-treated HRP cancer cells attracted PBMCs more abundantly, suggesting the potential for recruiting immune cells to HRP cancer cells through senescence-mediated
immunological activation. Our findings suggest that PARP inhibitors recruit immune cells to HRP cancer cells, potentially activating immune responses in the tumor microenvironment, providing
new insights into the clinical benefits of PARP inhibitors in immunotherapy for patients with HRP OC. SIMILAR CONTENT BEING VIEWED BY OTHERS CDC7 INHIBITION INDUCES REPLICATION
STRESS-MEDIATED ANEUPLOID CELLS WITH AN INFLAMMATORY PHENOTYPE SENSITIZING TUMORS TO IMMUNE CHECKPOINT BLOCKADE Article Open access 18 November 2023 THE STING PATHWAY: THERAPEUTIC
VULNERABILITIES IN OVARIAN CANCER Article 05 April 2022 PG545 SENSITIZES OVARIAN CANCER CELLS TO PARP INHIBITORS THROUGH MODULATION OF RAD51-DEK INTERACTION Article Open access 07 August
2023 INTRODUCTION Ovarian cancer (OC) is a fatal gynecological malignancy worldwide1. _BRCA1_ and _BRCA2_ are representative genes for homologous recombination (HR)-repair (HRR)2, and their
deleterious mutations are closely associated with hereditary and non-hereditary OC. Approximately 14–18% of patients with OC have germline mutations in _BRCA1_ or _BRCA2_, with an estimated
OC risk of 40–60% for _BRCA1_ and 11–27% for _BRCA2_3. These germline _BRCA1/2_ mutations also increase the risk of other cancers, including breast, prostate, and pancreatic cancers known as
hereditary breast and ovarian cancer (HBOC) syndrome4. Somatic mutations in _BRCA1/2_, on the contrary, are found in approximately 6% of OCs5. Beyond _BRCA1/2_, mutations in other
HRR-related genes, such as _RAD51_ and _PALB2_, are also detected in approximately 8% of OCs6. Overall, approximately 50% of advanced OCs are defined as HR-deficiency7. Owing to the high
prevalence of HR deficiency in OCs, the advent of poly (ADP-ribose) polymerase (PARP) inhibitors has revolutionized the therapeutic management of OC. PARP inhibitors target PARP enzymes,
primarily PARP1, which is essential for single-strand break (SSB) repair8. PARP1 binds to SSBs and activates and catalyzes poly (ADP-ribose) (PAR) synthesis to recruit DNA repair proteins8.
PARP inhibitors disrupt this process, leading to the accumulation of SSBs and subsequent double-strand breaks (DSBs) through replication fork collapse8. Certain PARP inhibitors also exert
the effects of “PARP trapping” on SSBs to generate DSBs more intensively9. DSBs are frequently repaired through HRR machineries; thus, HR-deficient (HRD) cancer cells exhibit higher
sensitivity to PARP inhibitors, in a process termed “synthetic lethality”8. Supporting their molecular mechanisms of action, several clinical trials have demonstrated significant
improvements in the prognosis of patients with primary advanced and recurrent HRD OC treated with PARP inhibitors7,10,11. The introduction of PARP inhibitors has contributed to increasing
the 5-year survival rate for OC from 36% in 1975 to 51% in 20191. However, the development of therapeutic strategies for HR-proficient (HRP) OC remains challenging. Various combination
treatments with PARP inhibitors are being evaluated for HRP cancers, including DNA damage response (DDR) inhibitors, anti-angiogenics, immune checkpoint inhibitors, chemotherapy, and
radiotherapy12. Notably, despite the limited efficacy of PARP inhibitors in HRP cancers in vitro13, several clinical trials have revealed some benefits for patients with HRP OC, although the
benefits have been inferior to those in HRD cases7,10. These findings indicate potential secondary antitumor mechanisms of PARP inhibitors beyond synthetic lethality. In this study, we
aimed to identify the role of PARP inhibitors in HRP OC patients. We found that PARP inhibitors induce senescence-like phenotypes in HRP cancer cells, upregulating a series of inflammatory
cytokine and chemokine genes, called the senescence-associated secretory phenotype (SASP). Furthermore, in vitro migration assays revealed that PARP inhibitor-treated HRP cancer cells
attracted peripheral blood mononuclear cells (PBMCs) more abundantly. Our findings suggest that PARP inhibitors recruit immune cells to HRP cancer cells, potentially activating immune
responses in the tumor microenvironment. MATERIALS AND METHODS COMPOUNDS Olaparib (AZD2281) and niraparib (MK-4827) were purchased from Selleck Chemicals (Houston, TX, USA). CELL LINES HeLa
and A549 cells were purchased from RIKEN BRC, and A549-reporter cells from InvivoGen (CA, USA). The STING knockout cells were generated in the previous study14. OVISE, TYK-nu, and MCAS cells
from JCRB Cell Bank, SKOV-3 cells from ATCC, and PBMCs from COSMO BIO CO., LTD. (Tokyo, Japan). Cells were cultured in DMEM or RPMI-1640 with 10% fetal bovine serum (FBS) and 1% penicillin
under standard conditions (37℃, 5% CO2). HRP cancer cells were defined as those without alterations in key HRR-associated genes15. IMMUNOFLUORESCENCE Immunofluorescence was performed as
previously described14. Anti-γH2A.X (Ser139) (20E3) (#9718S; Cell Signaling Technology, Danvers, MA, USA), anti-Lamin B1 (#66095–1-Ig; Proteintech, CH, USA) and anti-cGAS (D1D3G) (#15102;
Cell Signaling Technology) were used at 10 μg/mL. Images and cell counts were obtained using a BZ-X810 Analyzer (KEYENCE Corp., Osaka, Japan). Cells with five or more γH2A.X foci were
considered positive. CELLTITER-GLO LUMINESCENT CELL VIABILITY ASSAY Cell viability was assessed as previously described14 using the CellTiter-Glo™ 2.0 luminescent assay (Promega Corp.,
Madison, WI, USA). Luminescence was measured using SpectraMax Paradigm (Molecular Devices, LLC, San Jose, CA, USA). RNA PREPARATION AND QUANTITATIVE REVERSE TRANSCRIPTION-PCR (QRT-PCR) RNA
preparation and qRT-PCR were performed as previously reported14. Gene expression was normalized to GAPDH. TaqMan probe product IDs were as follows: human GAPDH (Hs99999905_m1), BRCA2
(Hs00609073_m1), IL1B (Hs01555410_m1), IL6 (Hs00174131_m1), CXCL10 (Hs00171042_m1), IFNB1 (Hs00277188_s1), and TNFSF15 (Hs00270802_s1). IMMUNOBLOTTING Immunoblotting followed previously
reported protocols14. Primary antibodies (1:1000 dilution) included anti-BRCA2 (D9S6V) (#10741; Cell Signaling Technology), anti-Rad51 (#ab63801; Abcam, Cambridge, UK), and anti-GAPDH
(14C10; #2118; Cell Signaling Technology). Proteins were visualized using ECL Select Detection Reagent (Cytiva, Marlborough, MA, USA), and the luminescent images were captured with
ImageQuant LAS 4010 (Cytiva, Marlborough, MA, USA). We processed images of blots according to the digital image and integrity policies of Scientific Reports. SIRNA KNOCKDOWN EXPERIMENT siRNA
knockdown was performed as previously described16. Human BRCA2 siRNA-SMARTpool (#M-003462–01–0005; Horizon) and Negative Control siRNA (#1022076; Qiagen, Hilden, Germany) were used. The
knockdown efficiency was evaluated using qRT-PCR and immunoblotting. DUAL REPORTER ASSAY A549-reporter cells (5.0 × 103/well) were seeded in 96-well plates and treated with DMSO or olaparib
(0.01–10 μM) for 120 h. Luciferase activity was measured with QUANTI-Luc (InvivoGen, #rep-qlc) and normalized to cell viability assessed by crystal violet staining as previously reported17.
Luminescence was measured using a SpectraMax Paradigm (Molecular Devices). TRANSCRIPTOME ANALYSIS HeLa cells (1.0 × 105/dish) were seeded on 10 cm dishes and treated with 10 μM DMSO or
olaparib for 72 h. Total RNA was extracted using the RNeasy kit (Qiagen), and libraries prepared by Rhelixa were sequenced on the Illumina platform. Data curation was performed as follows:
quality control (QC) checks by FastQC (version 0.11.9), trimming by trimmomatic (version 0.39), ribosomal RNA removal by bowtie (version 2.5.2), read mapping to a reference genome using STAR
(version 2.7.6), extraction of unique mapped reads by Samtools (version 1.19), and read counting using featureCounts (version 2.0.3). A comparative analysis using baySeq was performed in
the TCC library (version 1.40.0) using R (version 4.3.3). Enrichment analysis was performed using gene set enrichment analysis (GSEA) tools. SENESCENCE-ASSOCIATED Β-GALACTOSIDASE (SA-Β-GAL)
ACTIVITY ASSAY Cells (1.0 × 104/well) were seeded in 6-well plates and treated with 10 μM DMSO or PARP inhibitors (olaparib or niraparib) for 72 h. SA-β-GAL staining was performed at pH 6.0
using a Senescence β-Galactosidase Staining Kit (Cell Signaling Technology). Images were captured using a BZ-X810 Analyzer (KEYENCE Corp.), and SA-β-GAL-positive cells were manually counted.
FLUORESCENCE-ACTIVATED CELL SORTING (FACS) ANALYSIS Cells were fixed with 70% ethanol, washed with phosphate-buffered saline (PBS) containing 4% FBS, and stained with 20 μg/mL propidium
iodide (Thermo Fisher Scientific, Waltham, MA, USA) and 100 μg/mL RNase A (Nippon Gene, Tokyo, Japan). Ten thousand cells were analyzed using a FACSCanto™ II flow cytometer
(Becton–Dickinson, Franklin Lakes, NJ, USA). FlowJo software (v.10.7.1) was used for data analysis. PBMC MIGRATION ASSAY PBMC migration was assessed as previously described14. Conditioned
medium from cells treated with 10 μM DMSO or olaparib for 72 h was used. After 4 h incubation, migratory PBMCs fixed to the membrane were stained with crystal violet. Images were captured
using a BZ-9000 Analyzer (Keyence Corp.). STATISTICAL ANALYSIS Statistical analyses were performed using Excel (Microsoft Office) and GraphPad Prism (version 10.0.3). A two-sided Student’s
_t_ test and two-way ANOVA with Sidak’s multiple comparisons test were applied (*_P_ < 0.05, **_P_ < 0.01, ****_P_ < 0.0001; n.s., not significant). RESULTS EFFECT OF PARP
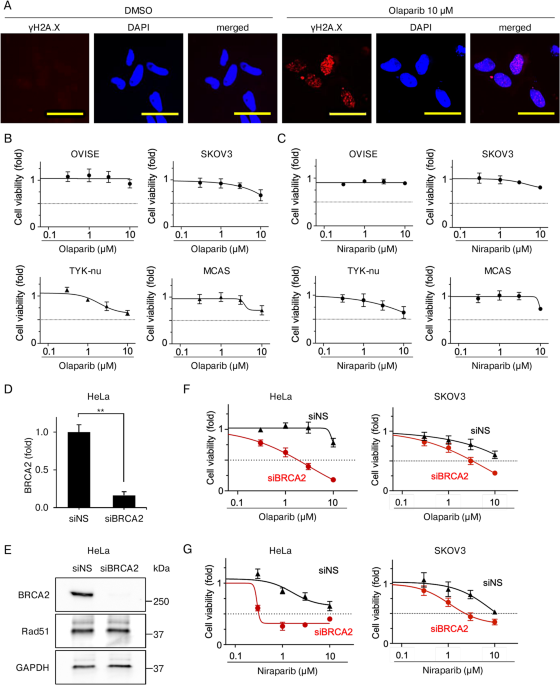
INHIBITORS ON CELL VIABILITY IN HRP CANCER CELLS IS LIMITED We determined the biological effects of PARP inhibitors on HRP OC cells in vitro. We first investigated the DNA-damaging activity
of olaparib in HeLa cells, selected for their suitability in various in vitro assays14,18. γH2 A.X foci, an index of DNA damage, was measured. Based on human pharmacokinetic data from
clinical trials19, the treatment of olaparib was set at 10 μM or lower. Immunofluorescence revealed a substantial increase in γH2 A.X foci-positive cells treated with olaparib for 72 h
compared with DMSO-treated controls (Fig. 1A, Supplementary Fig. 1, and Supplementary Table 1). These results indicate that olaparib caused DNA damage, even in HRP cancer cells. Next, the
antiproliferative activities of PARP inhibitors were determined in four HRP OC cell lines (OVISE, SKOV3, TYK-nu, and MCAS). Using CellTiter-Glo, cell viability was assessed 48 h
post-treatment. Minimal growth inhibition was observed in PARP inhibitor-treated cells at concentrations up to 1 μM, and none of them reached a survival rate below 50%, even at 10 μM (Fig.
1B–C). To confirm the contribution of HR proficiency to these outcomes, we knocked down _BRCA2_ in HeLa and SKOV3 cells using siRNA (siBRCA2). qRT-PCR revealed that _BRCA2_ siRNA achieved
84% knockdown (Fig. 1D), and western blotting confirmed effective inhibition of BRCA2 protein expression in BRCA2-depleted HeLa cells (Fig. 1E and Supplementary Fig. 2). CellTiter-Glo assays
in BRCA2-depleted HeLa and SKOV3 cells (HRD cells) treated with PARP inhibitors for 72 h revealed significant growth inhibition at ≥ 0.3 μM. Survival rates in BRCA2-depleted HeLa cells
dropped below 50% at ≥ 3 μM for olaparib and ≥ 1 μM for niraparib, while in BRCA2-depleted SKOV3 cells, a similar reduction was observed at ≥ 3 μM for both olaparib and niraparib (Fig.
1F–G). These results suggest that BRCA2 depletion markedly increases PARP inhibitor sensitivity in HRP cancer cells. Collectively, these results indicated that while PARP inhibitors could
induce DNA damage, their antiproliferative activities in HRP cancer cells were limited, suggesting a divergence of the biological effects of PARP inhibitors between DNA damage and
antiproliferation in HRP cancer cells. INFLAMMATORY CYTOKINE-RELATED PATHWAYS ARE TRANSCRIPTIONALLY UPREGULATED IN OLAPARIB-TREATED HELA CELLS To explore the mechanisms underlying PARP
inhibitors’ effects in HRP cancer cells, transcriptome analyses were performed in comparison between olaparib- and DMSO-treated HeLa cells. Cells were treated with 10 μM olaparib or DMSO for
72 h, where moderate anti-proliferation with considerable DNA damage was observed, and were subjected to RNA sequencing (RNA-seq) (Fig. 2A). In total, 17,118 and 16,786 transcripts were
detected in olaparib- and DMSO-treated cells, respectively. Analysis of differentially expressed genes (DEGs) revealed 866 genes, including 503 upregulated and 363 downregulated genes in
olaparib-treated cells (Fig. 2B–C and Supplementary Table 2–3). Volcano plots indicated the upregulation of inflammatory cytokine-related genes (Fig. 2C). Enrichment analyses using the Kyoto
Encyclopedia of Genes and Genomes (KEGG) database20,21 identified inflammation-related pathways as prominently upregulated in olaparib-treated cells, with five of the top 10 pathways
involving inflammation-related genes (Fig. 2D). Dot plot analyses also confirmed the upregulation of inflammatory hallmarks, such as TNFA signaling via NFκB, inflammatory response, IL2-STAT5
signaling and interferon gamma response, in olaparib-treated cells (Fig. 2E). Furthermore, the GSEA determined significant enrichment of inflammatory cytokine-related KEGG pathways,
including cytokine–cytokine receptor interaction, JAK-STAT signaling, and Rig I like receptor signaling pathways, as well as inflammatory hallmarks such as TNFA signaling via NFκB,
interferon alfa response, and interferon gamma response in olaparib-treated cells (Fig. 2F). To confirm the upregulation of inflammatory response genes, reporter assays were performed in
A549-based interferon regulatory factor (IRF)-Lucia luciferase (Luc) reporter cells (A549 reporter cells). Treatment with olaparib for 120 h showed significant dose-dependent increases in
IRF-Luc activities, 2- and 3-fold with olaparib treatment at 1 μM and 10 μM, respectively (Fig. 2G, red bars), whereas the antiproliferative activities were limited, with survival rates
above 50% (Fig. 2G, black line). Collectively, our findings demonstrate that PARP inhibitors upregulate inflammation-related pathways in HRP cancer cells despite limited antiproliferative
activities. PARP INHIBITORS INDUCE CELLULAR SENESCENCE IN HRP CANCER CELLS Considering the close correlation between DNA damage-induced inflammatory activation and a senescence-like
phenotype, termed SASP22, we next determined whether olaparib-treated HRP cancer cells exhibited senescence-like phenotypes based on morphological alteration, SA-β-GAL activity, and cellular
granularity in seven cell lines: four OC cell lines (OVISE, SKOV3, TYK-nu, and MCAS) and three other cancer cell lines (HeLa, A549, and A549-reporter). The olaparib-treated cells flattened
and enlarged 24 h after treatment, and their cytomegalic phenotype progressed until 72 h, with enlarged nuclei or grape-like multinuclear clusters, which are hallmarks of DNA damage-induced
senescence-like cells23 (Fig. 3A and Supplementary Fig. 3). To further assess the senescence-like phenotypes, we determined SA-β-GAL activity and cytoplasmic granules, which are common
senescence markers23,24. The number of SA-β-GAL-positive cells considerably increased in all seven cell lines treated with olaparib compared to controls (Fig. 3B and Supplementary Table 4).
Similar to olaparib treatment, niraparib also increased SA-β-GAL activity in HeLa, OVISE, and SKOV3 cells, along with morphological alterations of senescence-like phenotype (Fig. 3C–D and
Supplementary Table 4). Next, we identified cytoplasmic granules in olaparib-treated cells using FACS. Cellular granularity is evaluated using side-scatter (SSC) values, reflecting
intracellular complexity, compared to forward-scatter (FSC) values24. In HeLa cells, treatment with 10 μM olaparib for 72 h increased both SSC and FSC (Fig. 3E, Q2: 6.57% in olaparib and
2.92% in DMSO), with SSC increase being more substantially increased (Fig. 3E, Q1: 8.07% in olaparib and 1.03% in DMSO), indicating an enlarged morphology with intracellular granular
accumulation. Similar increases in SSC were also observed in the niraparib-treated HeLa cells (Fig. 3F). Furthermore, elevated SSC values were observed in olaparib-treated A549,
A549-reporter and MCAS cells, as well as in the niraparib-treated SKOV3 cells (Fig. 3G–H). These results suggest that PARP inhibitor-induced DNA damage causes cellular senescence or
senescence-like phenotypes, with elevated inflammatory gene expression in HRP cancer cells, despite limited antiproliferative effects. CGAS-STING SIGNALING PATHWAY IS INVOLVED IN CELLULAR
SENESCENCE INDUCED BY THE PARP INHIBITOR IN HRP CANCER CELLS Senescent cells upregulate the expression of inflammatory cytokines and chemokines, termed SASP22. Next, we determined the
SASP-related gene expression in olaparib-treated HRP cancer cells. _IL-1B_, a representative SASP factor, was significantly upregulated in a dose-dependent manner in olaparib-treated HeLa
cells (by 2.7-fold at 3 μM, 19.3-fold at 10 μM, and 77.1-fold at 30 μM; Fig. 4A) and A549-reporter cells (by 2.3-fold at 1 μM, 17.9-fold at 3 μM, 17.1-fold at 10 μM, and 59.5-fold at 30 μM;
Fig. 4B). Other SASP factors, _IL-6_ and _CXCL10_, also increased after treatment with olaparib at concentrations ranging from 3 μM to 30 μM in both HeLa and A549-reporter cells (Fig. 4A–B).
In three HRP OC cells (OVISE, SKOV3, and MCAS) and A549 cells, _IL1B_ expression also significantly increased in a dose-dependent manner after olaparib treatment (Fig. 4C). In addition,
_IFNB1_ and _TNFSF15_, which are representative genes in type-I interferon and TNF-α pathways that activate cellular senescence25,26, were also significantly upregulated in olaparib-treated
OVISE, SKOV3, and MCAS cell lines (Fig. 4D). Multiple studies have demonstrated the involvement of cGAS-STING pathway in PARP inhibitor-associated inflammation27,28,29. Immunofluorescence
revealed that olaparib treatment profoundly generated micronuclei in both A549 and SKOV3 cells, where cGAS protein was intensively localized (Fig. 4E). Furthermore, knockout of STING
drastically suppressed the reporter activity of Luc-IRF in olaparib-treated A549-reporter cells (Fig. 4F), indicating that the cGAS-STING pathway plays critical roles in inflammatory
responses in the PARP inhibitor-treated cells. SASP PRODUCED IN OLAPARIB-TREATED HRP CANCER CELLS RECRUITS PBMCS IN VITRO MIGRATION ASSAYS Finally, we determined whether the PARP
inhibitor-induced senescence-like cells could attract PBMCs via secretory factors. In vitro migration assays were performed with PBMCs and conditioned medium from olaparib-treated HRP cancer
cells, using HeLa and MCAS cells treated with 10 μM DMSO or olaparib for 72 h (Fig. 5A). The chemoattractant activities were assessed by quantifying PBMCs that migrated through the
membrane, stained with crystal violet. The number of PBMCs on the membrane profoundly increased in the conditioned medium from olaparib-treated cells compared to that from controls in both
HeLa and MCAS cell lines (Fig. 5B), suggesting that olaparib-treated HRP cancer cells could immunologically activate PBMCs through secreted chemoattractants. Collectively, our findings
demonstrate that PARP inhibitors induce cellular senescence with less cell death activity in HRP cancer cells, potentially leading to immunological activation to attract blood cells to HRP
cancer cells (Fig. 5C). DISCUSSION In this study, we demonstrated that PARP inhibitors induce senescence-like phenotypes to produce SASP factors in HRP cancer cells, although their
antiproliferative effects are limited. In vitro migration assays confirmed that conditioned medium from PARP inhibitor-treated HRP cells attracted PBMCs more profoundly than conditioned
medium from control-treated cells. These findings suggest that PARP inhibitors enhance immune cell recruitment to HRP cancer cells through senescence-mediated immunological activation. PARP
inhibitors have dramatically improved the poor prognosis of OC owing to the high prevalence of HRD in advanced cases, known as “synthetic lethality.” In contrast, treatment strategies for
HRP OC have not been fully established yet. In some clinical trials, unexpectedly, PARP inhibitors exhibited marginal efficacy even in patients with HRP OC7,10, hinting at potential
antitumor activity that is mechanistically distinct from cytotoxicity by “synthetic lethality.” Recent studies have demonstrated that PARP inhibitors induce cellular senescence through the
accumulation of DNA damage30,31. The p53/p21 and p16/RB pathways are involved in senescence-associated antiproliferative effects22. Furthermore, even in p53-mutant OC cell lines, PARP
inhibitors induce p53-independent but p21-mediated cellular senescence accompanied by DNA damage response (DDR) activation30, suggesting diverse mechanisms underlying cellular senescence in
cancer cells. We found that PARP inhibitors induced senescence-like phenotypes in OC and other HRP cancer cells, which was confirmed using various senescence biomarkers: morphological
alterations, SA-β-GAL activity, cellular granularity, and SASP-related gene expression (Fig. 3A–H, Fig. 4A–D). Transcriptome analyses revealed a robust upregulation of gene sets and pathways
associated with inflammatory cytokines and chemokines in PARP inhibitor-treated HRP cancer cells, strongly indicating senescence induction in HRP OC and other cancer types. The effects of
senescence on tumor development, including tumor-promoting or tumor-suppressive effects, have been reported to be controversial. Senescent cells produce secretory proteins termed SASP
factors, including inflammatory cytokines, chemokines, growth factors, and MMP22,32. SASP factors facilitate tumor progression by promoting tumor growth, epithelial–mesenchymal transition
(EMT), and immunosuppression32. Based on the concepts of “tumor-promotive effects of senescence,” novel therapeutic strategies of senolytics, which eliminates senescent cells, or
senomorphics, which blocks SASP factors, are under development32. Conversely, SASP factors also contribute to tumor regression by activating immune surveillance, enhancing the infiltration
of natural killer (NK) cells, macrophages, and T cells, and facilitating the clearance of senescent tumor cells32. In our study, PARP inhibitor-treated HeLa cells showed a significant
increase in the expression of _CXCL10_ and other chemokines (Fig. 4A), which promoted T-cell infiltration33. KEGG-based enrichment analysis of DEGs revealed that NK cell-mediated
cytotoxicity pathways were significantly enriched in the PARP inhibitor-treated cells (Fig. 2D). In addition to these senescent phenotypes and gene expression profiles, conditioned medium
from PARP inhibitor-treated HRP cancer cells profoundly attracted PBMCs in the in vitro migration assays (Fig. 5B). Collectively, PARP inhibitors are expected to boost immunological
activation in the TME by recruiting blood cells via the induction of senescence in cancer cells. These inflammatory effects of PARP inhibitors may contribute to the antitumor efficacy of HRP
in OC. In conclusion, we demonstrated that PARP inhibitors induce cellular senescence in HRP and other cancer cells, upregulating inflammatory cytokines and chemokines to attract PBMCs in
vitro. These findings suggest the potential for recruiting immune cells to HRP cancer cells through senescence-mediated immunological activation. Further studies will provide new insights
into the clinical benefits of PARP inhibitors with immunotherapy for patients with HRP OC. DATA AVAILABILITY The datasets used and/or analyzed during the current study available from the
corresponding author on reasonable request. REFERENCES * Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. _CA Cancer J. Clin._ 74, 12–49.
https://doi.org/10.3322/caac.21820 (2024). Article PubMed Google Scholar * Yoshida, K. & Miki, Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in
response to DNA damage. _Cancer Sci._ 95, 866–871. https://doi.org/10.1111/j.1349-7006.2004.tb02195.x (2004). Article CAS PubMed Google Scholar * Lheureux, S., Braunstein, M. & Oza,
A. M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. _CA: A Cancer J. Clin._ https://doi.org/10.3322/caac.21559 (2019). Article Google Scholar *
Daly, M. B. et al. NCCN Guidelines® Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2024. _J. Natl. Compr. Cancer Netw._ 21, 1000–1010.
https://doi.org/10.6004/jnccn.2023.0051 (2023). Article CAS Google Scholar * Arcieri, M. et al. How BRCA and homologous recombination deficiency change therapeutic strategies in ovarian
cancer: A review of literature. _Front. Oncol._ 14, 1335196. https://doi.org/10.3389/fonc.2024.1335196 (2024). Article CAS PubMed PubMed Central Google Scholar * Pennington, K. P. et
al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. _Clin. Cancer Res._ 20,
764–775. https://doi.org/10.1158/1078-0432.CCR-13-2287 (2014). Article CAS PubMed Google Scholar * González-Martín, A. et al. Niraparib in patients with newly diagnosed advanced ovarian
cancer. _N. Engl. J. Med._ 381, 2391–2402. https://doi.org/10.1056/nejmoa1910962 (2019). Article CAS PubMed Google Scholar * Lord, C. J. & Ashworth, A. PARP inhibitors: Synthetic
lethality in the clinic. _Science_ 355, 1152–1158. https://doi.org/10.1126/science.aam7344 (2017). Article ADS CAS PubMed PubMed Central Google Scholar * Murai, J. et al. Trapping of
PARP1 and PARP2 by clinical PARP inhibitors. _Cancer Res._ 72, 5588–5599. https://doi.org/10.1158/0008-5472.CAN-12-2753 (2012). Article CAS PubMed PubMed Central Google Scholar * Monk,
B. J. et al. A randomized, phase III trial to evaluate rucaparib monotherapy as maintenance treatment in patients with newly diagnosed ovarian cancer (ATHENA–MONO/GOG-3020/ENGOT-ov45). _J.
Clin. Oncol._ 40, 3952–3964. https://doi.org/10.1200/jco.22.01003 (2022). Article CAS PubMed PubMed Central Google Scholar * Pujade-Lauraine, E. et al. Olaparib tablets as maintenance
therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. _Lancet Oncol._
18, 1274–1284. https://doi.org/10.1016/S1470-2045(17)30469-2 (2017). Article CAS PubMed Google Scholar * Xie, Y. et al. Combined strategies with PARP inhibitors for the treatment of
BRCA wide type cancer. _Front. Oncol._ https://doi.org/10.3389/fonc.2024.1441222 (2024). Article PubMed PubMed Central Google Scholar * Farmer, H. et al. Targeting the DNA repair defect
in BRCA mutant cells as a therapeutic strategy. _Nature_ 434, 917–921. https://doi.org/10.1038/nature03445 (2005). Article ADS CAS PubMed Google Scholar * Morita, T. Y. et al. CDC7
inhibition induces replication stress-mediated aneuploid cells with an inflammatory phenotype sensitizing tumors to immune checkpoint blockade. _Nat. Commun._ 14, 7490.
https://doi.org/10.1038/s41467-023-43274-3 (2023). Article ADS CAS PubMed PubMed Central Google Scholar * De Bono, J. et al. Olaparib for metastatic castration-resistant prostate
cancer. _N. Engl. J. Med._ 382, 2091–2102. https://doi.org/10.1056/nejmoa1911440 (2020). Article CAS PubMed Google Scholar * Yamamoto, G. et al. WEE1 confers resistance to KRAS(G12C)
inhibitors in non-small cell lung cancer. _Cancer Lett._ 611, 217414. https://doi.org/10.1016/j.canlet.2024.217414 (2024). Article CAS PubMed Google Scholar * Fujimoto, Y. et al.
Combination treatment with a PI3K/Akt/mTOR pathway inhibitor overcomes resistance to anti-HER2 therapy in PIK3CA-mutant HER2-positive breast cancer cells. _Sci. Rep._ 10, 21762.
https://doi.org/10.1038/s41598-020-78646-y (2020). Article ADS CAS PubMed PubMed Central Google Scholar * Iwai, K. et al. A CDC7 inhibitor sensitizes DNA-damaging chemotherapies by
suppressing homologous recombination repair to delay DNA damage recovery. _Sci. Adv._ https://doi.org/10.1126/sciadv.abf0197 (2021). Article PubMed PubMed Central Google Scholar *
Yonemori, K. et al. Safety and tolerability of the olaparib tablet formulation in Japanese patients with advanced solid tumours. _Cancer Chemother. Pharmacol._ 78, 525–531.
https://doi.org/10.1007/s00280-016-3106-7 (2016). Article CAS PubMed PubMed Central Google Scholar * Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. _Nucleic Acids Res._ 28,
27–30. https://doi.org/10.1093/nar/28.1.27 (2000). Article CAS PubMed PubMed Central Google Scholar * Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M.
KEGG: biological systems database as a model of the real world. _Nucleic Acids Res._ 53, D672–D677. https://doi.org/10.1093/nar/gkae909 (2025). Article PubMed Google Scholar * Coppé,
J.-P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. _PLoS Biol._ 6, e301.
https://doi.org/10.1371/journal.pbio.0060301 (2008). Article CAS PubMed PubMed Central Google Scholar * Huang, W., Hickson, L. J., Eirin, A., Kirkland, J. L. & Lerman, L. O.
Cellular senescence: The good, the bad and the unknown. _Nat. Rev. Nephrol._ 18, 611–627. https://doi.org/10.1038/s41581-022-00601-z (2022). Article CAS PubMed PubMed Central Google
Scholar * Hwang, E. S., Yoon, G. & Kang, H. T. A comparative analysis of the cell biology of senescence and aging. _Cell. Mol. Life Sci._ 66, 2503–2524.
https://doi.org/10.1007/s00018-009-0034-2 (2009). Article CAS PubMed PubMed Central Google Scholar * Frank, T. et al. Cell cycle arrest in mitosis promotes interferon-induced
necroptosis. _Cell Death Differ._ 26, 2046–2060. https://doi.org/10.1038/s41418-019-0298-5 (2019). Article PubMed PubMed Central Google Scholar * Tanzer, M. C. A proteomic perspective on
TNF-mediated signalling and cell death. _Biochem. Soc. Trans._ 50, 13–20. https://doi.org/10.1042/bst20211114 (2022). Article CAS PubMed PubMed Central Google Scholar * Kim, C., Wang,
X. D. & Yu, Y. PARP1 inhibitors trigger innate immunity via PARP1 trapping-induced DNA damage response. _Elife_ 9, e60637. https://doi.org/10.7554/elife.60637 (2020). Article CAS
PubMed PubMed Central Google Scholar * Wang, T. et al. Combination of <scp>PARP</scp> inhibitor and <scp>CDK4</scp>/6 inhibitor modulates
<scp>cGAS</scp>/<scp>STING-</scp>dependent therapy-induced senescence and provides “one-two punch” opportunity with <scp>anti-PD-L1</scp> therapy in
colorectal cancer. _Cancer Sci._ 114, 4184–4201. https://doi.org/10.1111/cas.15961 (2023). Article CAS PubMed PubMed Central Google Scholar * Pantelidou, C. et al. STING agonism
enhances anti-tumor immune responses and therapeutic efficacy of PARP inhibition in BRCA-associated breast cancer. _NPJ Breast Cancer_ https://doi.org/10.1038/s41523-022-00471-5 (2022).
Article PubMed PubMed Central Google Scholar * Fleury, H. et al. Exploiting interconnected synthetic lethal interactions between PARP inhibition and cancer cell reversible senescence.
_Nat. Commun._ https://doi.org/10.1038/s41467-019-10460-1 (2019). Article PubMed PubMed Central Google Scholar * Lombard, A. P. et al. Olaparib-Induced senescence is bypassed through
G2–M checkpoint override in olaparib-resistant prostate cancer. _Mol. Cancer Ther._ 21, 677–685. https://doi.org/10.1158/1535-7163.mct-21-0604 (2022). Article CAS PubMed PubMed Central
Google Scholar * Dong, Z. et al. Cellular senescence and SASP in tumor progression and therapeutic opportunities. _Mol. Cancer_ https://doi.org/10.1186/s12943-024-02096-7 (2024). Article
PubMed PubMed Central Google Scholar * Muthuswamy, R. et al. NF-κB hyperactivation in tumor tissues allows tumor-selective reprogramming of the chemokine microenvironment to enhance the
recruitment of cytolytic T effector cells. _Can. Res._ 72, 3735–3743. https://doi.org/10.1158/0008-5472.can-11-4136 (2012). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS
We would like to thank members of the Aki Lab: Kumi Kinoshita, Emi Ito, Yasushi Kawagishi, Takuya Nakatsuru, and Keita Hirouchi. We also thank Katsuya Tsuchihara, Tohru Kiyono, Riu
Yamashita for their valuable comments on the manuscript and technical support. FUNDING This research was supported by the Grants-in-Aid for Scientific Research (B) (20H03541) to A.O., the
Grants-in-Aid for Fostering Joint International Research (B) (20KK0187) to A.O., Takeda Science Foundation (2019, 2022) to A.O., the Grants-in-Aid for Scientific Research (C) (23K06708) for
R.K., and the Grant-in-Aid for Research Activity Start-up (24K23370) for K.N. AUTHOR INFORMATION Author notes * Misato Kamii, Ryo Kamata contributed equally as co-first authors. AUTHORS AND
AFFILIATIONS * Division of Collaborative Research and Developments, Exploratory Oncology Research & Clinical Trial Center, National Cancer Center, 6-5-1 Kashiwanoha, Kashiwa, Chiba,
277-8577, Japan Misato Kamii, Ryo Kamata, Hitoshi Saito, Gaku Yamamoto, Chiaki Mashima, Toyohiro Yamauchi, Takehiro Nakao, Yuta Sakae, Tomoko Yamamori-Morita, Kazuki Nakai, Yumi Hakozaki
& Akihiro Ohashi * Department of Obstetrics and Gynecology, The Jikei University School of Medicine, 3-25-8 Nishi-Shimbashi, Minato-Ku, Tokyo, 105-8461, Japan Misato Kamii, Masataka
Takenaka & Aikou Okamoto * Department of Integrated Bioscience, Graduate School of Frontier Sciences, The University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa, Chiba, 277-0882, Japan Toyohiro
Yamauchi & Akihiro Ohashi Authors * Misato Kamii View author publications You can also search for this author inPubMed Google Scholar * Ryo Kamata View author publications You can also
search for this author inPubMed Google Scholar * Hitoshi Saito View author publications You can also search for this author inPubMed Google Scholar * Gaku Yamamoto View author publications
You can also search for this author inPubMed Google Scholar * Chiaki Mashima View author publications You can also search for this author inPubMed Google Scholar * Toyohiro Yamauchi View
author publications You can also search for this author inPubMed Google Scholar * Takehiro Nakao View author publications You can also search for this author inPubMed Google Scholar * Yuta
Sakae View author publications You can also search for this author inPubMed Google Scholar * Tomoko Yamamori-Morita View author publications You can also search for this author inPubMed
Google Scholar * Kazuki Nakai View author publications You can also search for this author inPubMed Google Scholar * Yumi Hakozaki View author publications You can also search for this
author inPubMed Google Scholar * Masataka Takenaka View author publications You can also search for this author inPubMed Google Scholar * Aikou Okamoto View author publications You can also
search for this author inPubMed Google Scholar * Akihiro Ohashi View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.K.: Data curation, Formal
analysis, Investigation, Visualization, Writing¬–original draft, R.K.: Conceptualization, Formal analysis, Investigation, Writing¬–review & editing H.S.: Formal analysis, Investigation,
G.Y.: Resources, C.M.: Resources, T.Y.: Resources, T.N.: Resources, Y.S.: Resources, T.Y.M.: Resources, K.N.: Resources, Y.H.: Resources, M.T.: Supervision, A.O.: Supervision, A.Oh.:
Conceptualization, Funding acquisition, Project administration, Supervision, Writing¬–original draft, Writing¬–review & editing. CORRESPONDING AUTHORS Correspondence to Ryo Kamata or
Akihiro Ohashi. ETHICS DECLARATIONS COMPETING INTERESTS T.Y. was an employee of Axcelead Drug Discovery Partners, Inc (2023-present). A.Oh. was an employee of Takeda Pharmaceutical Company,
Ltd (2006–2018). A.Oh. is a part-time employee of Astellas Pharma Inc. as a cross-appointment system (2022-present). A.Oh. reported paid consulting or advisory roles for Ono Pharmaceutical
Company Ltd., Craif Inc., and GEXVal Inc. out of this study. A.Oh. has received research funding from Takeda Pharmaceutical, Daiichi-Sankyo, and Astellas Pharma companies out of this study.
M.K., R.K., H.S., G.Y., C.M., T.N., Y.S., T.Y.M, K.N., Y.H., M.T., and A.O. declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with
regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is
licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any
medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed
material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are
included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and
your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this
licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kamii, M., Kamata, R., Saito, H. _et al._ PARP inhibitors
elicit a cellular senescence mediated inflammatory response in homologous recombination proficient cancer cells. _Sci Rep_ 15, 15458 (2025). https://doi.org/10.1038/s41598-025-00336-4
Download citation * Received: 10 January 2025 * Accepted: 28 April 2025 * Published: 02 May 2025 * DOI: https://doi.org/10.1038/s41598-025-00336-4 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative KEYWORDS * Poly (ADP-ribose) polymerase (PARP) inhibitors * Homologous recombination-proficient (HRP) cancer * Ovarian cancer (OC) *
Senescence-associated secretory phenotype (SASP)
