Task-specific odorant receptor expression in worker antennae indicates that sensory filters regulate division of labor in ants
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Division of labor (DOL) is a characteristic trait of insect societies, where tasks are generally performed by specialized individuals. Inside workers focus on brood or nest care,
while others take risks by foraging outside. Theory proposes that workers have different thresholds to perform certain tasks when confronted with task-related stimuli, leading to
specialization and consequently DOL. Workers are presumed to vary in their response to task-related cues rather than in how they perceive such information. Here, we test the hypothesis that
DOL instead stems from workers varying in their efficiency to detect stimuli of specific tasks. We use transcriptomics to measure mRNA expression levels in the antennae and brain of nurses
and foragers of the ant _Temnothorax longispinosus_. We find seven times as many genes to be differentially expressed between behavioral phenotypes in the antennae compared to the brain.
Moreover, half of all odorant receptors are differentially expressed, with an overrepresentation of the 9-exon gene family upregulated in the antennae of nurses. Nurses and foragers thus
apparently differ in the perception of their olfactory environment and task-related signals. Our study supports the hypothesis that antennal sensory filters predispose workers to specialize
in specific tasks. SIMILAR CONTENT BEING VIEWED BY OTHERS DUFOUR’S GLAND ANALYSIS REVEALS CASTE AND PHYSIOLOGY SPECIFIC SIGNALS IN _BOMBUS IMPATIEN_S Article Open access 02 February 2021
MICROSTRUCTURES AT THE DISTAL TIP OF ANT CHEMOSENSORY SENSILLA Article Open access 11 November 2022 BUMBLE BEE QUEEN PHEROMONES ARE CONTEXT-DEPENDENT Article Open access 20 August 2021
INTRODUCTION Division of labor (DOL) is an important organizing principle of complex biological systems that arose independently during three of the major evolutionary transitions1. DOL was
originally formulated in the context of the production process in human societies2, but specialization to specific tasks is also found within cells and across many organisms1. Examples range
from bacteria, where clonal populations are divided into subpopulations focusing on different activities3,4,5,6 to multicellular organisms with differentiation of cells into different
tissues and organs, and individuals performing specific roles in animal societies7,8. To understand the evolution of complex life, it is therefore essential to investigate the mechanisms
that underlie DOL. DOL in insect societies results from individuals specializing in the performance of specific tasks. In addition to the reproductive DOL between fertile queens and
functionally sterile workers, there is a behavioral DOL among workers that specialize in tasks such as brood care, foraging, nest building, and defense9,10,11. Several factors can affect
task specialization, including age12,13, nutrition14,15, morphology16, genotype17,18,19, experience20, and colony size21,22,23. In leafcutter ant species, among others, behavioral
specialization of morphologically distinct groups of workers contributes to the DOL24. In most social insect species, younger workers tend to perform intranidal tasks, while older
individuals perform risky activities such as nest defense and foraging outside the nest25,26. Yet, such specialization among workers remains flexible, as foragers can revert to perform brood
care when needed27,28. Several molecular mechanisms have been implicated in the regulation of DOL. Task specialization is associated with transcriptional changes in the worker
brain29,30,31,32,33,34. Molecular pathways such as the insulin/insulin-like signaling (IIS), vitellogenin (Vg), and juvenile hormone (JH) pathways are involved in the regulation of worker
behavior35,36,37,38,39,40. Functional manipulations have confirmed that the expression of key genes in the worker brain controls task specialization41,42,43. Behavioral variation among
workers is also associated with signaling of biogenic amines (e.g., dopamine, octopamine, tyramine, serotonin), which act as neurotransmitters or neuromodulators involved in the modulation
of the responsiveness to task-associated stimuli44,45,46,47. Self-organization and collective behavior in insect societies are maintained via the exchange of chemical information48. Social
insects communicate primarily through glandular pheromones and complex mixtures of long-chain hydrocarbons on their cuticle. These cuticular hydrocarbons (CHC) facilitate recognition of
nestmates, developmental stages, castes, sexes, and species49,50. Social insects perceive chemical information via different types of sensilla in the antennae, small receptor organs embedded
in the integument that are connected to sensory neurons51,52,53. Decoding the identities of chemical compounds relies on odorant receptors (ORs) located within each sensillum54,55. ORs are
transmembrane proteins expressed in the dendrites of olfactory receptor neurons (ORN). The largely conserved OR coreceptor (_Orco_) of insects56 is required for odorant recognition in the
dendritic membrane: it forms an ion channel with specific OR, which determines the sensitivity and specificity of the ORN57. Odorant molecules penetrate through the antenna cuticular pores
and are transported by odorant-binding proteins to the ORN membrane, where they interact with receptors, leading to the generation of action potentials58,59. ORN axons relay signals from the
sensilla to the glomeruli of the antennal lobes in the insect brain, which are the first processing unit for olfactory information. Then, ORN make synaptic contact with the projection
neurons and local neurons, which transfer information to the central brain60,61,62. Several lines of evidence indicate that OR genes play central roles in the regulation of social life of
insects. First, social insect species typically harbor large numbers of OR genes63,64,65,66,67. Second, some OR gene families have specifically expanded during social evolution, such as the
9-exon subfamily in ants, which appears to serve an important function in the perception of CHC63,66,68,69,70,71. Third, species that evolved socially parasitic strategies resulting in
reduced behavioral repertoires show a strong and convergent reduction in the number of OR genes72. Finally, experimentally produced mutant ants that lack the _Orco_ gene (coding for the
co-receptor necessary for OR to properly function) show impaired social behavior73,74. Current DOL models bring together the chemical nature of social insect communication and variation
among workers in their response to chemical cues. They posit that flexible response thresholds to task-related chemical stimuli serve as regulators of worker specialization75. Workers take
on a particular task when the stimulus intensity exceeds their individual threshold for this task. Therefore, individuals with lower thresholds for a given task are more likely to perform it
than those with higher thresholds76. Individual response decisions and task performance are modulated via numerous parameters on different time scales7,77,78,79, and despite extensive
research on DOL and task allocation, these mechanisms are not fully understood. As the name suggests, response threshold models are based on an individual’s response to certain stimuli and
the variation in their responsiveness over time. However, these studies do not include in their models how these cues are processed65,79,80,81,82,83,84,85. The fact that the processing of
signal information has been primarily described in the insect brain (see ref. 86) may suggest that thresholds and associated responses are set in the central nervous system, possibly
regulated via molecular pathways in the brain that correlate with behavioral variation. We propose that DOL models would benefit from considering odor sensitivity as a potential upstream
sensory filter that may affect task specialization. Along these lines, we hypothesize that behavioral variation among workers may also stem from their ability and/or efficiency in detecting
different signals. For example, we propose that individuals that specialize in brood care do so because they are more sensitive to brood cues, rather than (or in addition to) being more
likely to respond to similar levels of brood cues. To test the hypothesis that a sensory filter regulates inter-individual behavioral variation, and thus the DOL in social insects, we
investigated transcriptional signals in the brain and antennae of workers that specialize in either brood care or foraging behavior in the ant _Temnothorax longispinosus_. We found that (i)
behavioral variation was associated with more extensive transcriptomic differences in the antennae than in the brain, (ii) these differences included a large proportion of the OR gene
repertoire, and (iii) individuals specializing in brood care overexpressed an OR gene family putatively involved in detecting social cues. These findings support our hypothesis that the
peripheral nervous system, acting as a sensory filter, plays an important role in regulating behavioral differences between workers and thus in the DOL of social insects. RESULTS
IDENTIFICATION OF _TEMNOTHORAX LONGISPINOSUS_ BEHAVIORAL PHENOTYPES Task specialization in the ant _T. longispinosus_ is neither genetically fixed nor rigid, but can change with age and in
response to colony needs39. We conducted behavioral observations of seven _T. longispinosus_ laboratory colonies to identify individuals that specialize in brood care behavior (hereafter
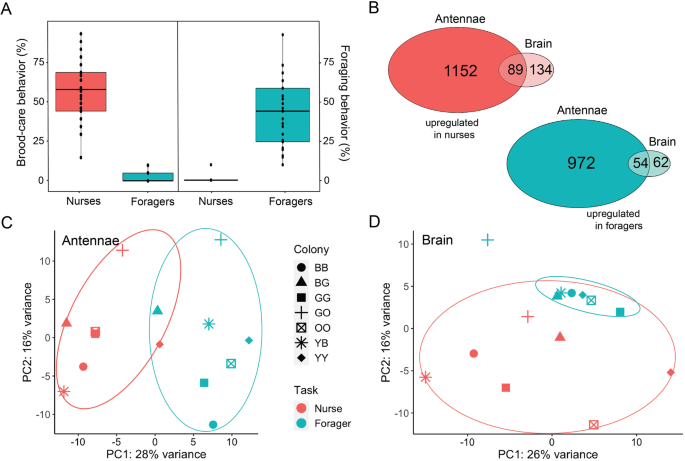
referred to as nurses), and others that specialize in foraging (hereafter referred to as foragers) (Fig. 1A; see “Methods” section for more details). The grouping of workers into the two
behavioral categories was based on the frequency of their location inside or outside the colony and on their behavior, especially whether they showed brood care or foraging behavior (Dataset
S1). Individuals identified as nurses interacted with the brood in 54% ± 22% (mean ± sd) of the observations, and were recorded outside the nest in 1% ± 3% of the observations. On the
contrary, foragers were found outside the nest in 42% ± 26% of the observations but interacted with the brood in only 2% ± 4% of the observations. LARGER TASK-ASSOCIATED TRANSCRIPTOMIC
CHANGES IN THE ANTENNAE THAN IN THE BRAIN To investigate transcriptomic variation between nurses and foragers, we used RNA-seq to generate seven nurse and seven forager brain and antenna
samples, each consisting of pooled tissue from seven workers of a single colony. Of the 14,837 genes annotated in the _T. longispinosus_ genome, we found 91% (13,494) and 92% (13,683) to be
expressed (FPKM > 0) in the brain and the antennae, respectively. To investigate gene expression differences between nurses and foragers, we compared full models that included task as an
explanatory variable to reduced models that did not using the likelihood ratio test (LRT) method implemented in DEseq2. The influence of colony identity was controlled for by including it as
an explanatory variable in both full and reduced models. A Benjamini–Hochberg adjusted _p_ value of 0.05 was set as a threshold to obtain genes whose variation was significantly explained
by behavioral specialization in each tissue. We detected 339 differentially expressed genes (DEGs) in the brain (223 upregulated in nurses, 116 in foragers), and 2267 in the antennae (1241
upregulated in nurses, 1026 in foragers; Dataset S2 and Fig. 1B). We found an overlap of 162 DEGs between the two tissues, including 143 DEGs that showed differences in the same direction
across tissues. Principal component analyses (PCA) for both brain and antenna data reveal that most samples clustered by behavioral phenotype rather than colony of origin (Fig. 1C, D). OR
GENE EXPRESSION DIFFERS BETWEEN ANTENNAE OF NURSES AND FORAGERS To investigate whether odorant perception differs between nurses and foragers, we focused our attention on the expression of
OR genes in the antennae. We found that all 419 previously annotated OR genes in the genome of _T. longispinosus_72 were expressed in the antennae, and that 50% (209/419) of them were
differentially expressed between nurses and foragers. Specifically, 64 OR genes were upregulated in nurses (15% of all OR genes), and 145 in foragers (35% of all ORs) (Fig. 2). Then, we
studied which OR subfamilies were preferentially expressed in nurses and foragers. The 64 OR genes overexpressed in nurses belonged to three OR subfamilies, while the 145 OR genes
upregulated in foragers were distributed among 19 OR subfamilies. Foragers overexpressed 27, 19, 8, and 8 OR genes from the L, V, P, and H subfamilies, respectively, while no OR genes from
these subfamilies were upregulated in nurses (Dataset S3). We found that 63% (27/43) of the genes from the L subfamily and 73% (8/11) from the P subfamily were overexpressed in foragers,
which represents a significant overrepresentations (L subfamily: Fisher’s test, odds ratio = 3.67, _p_ value < 0.001; P subfamily: Fisher’s test, odds ratio = 5.25, _p_ value < 0.02).
For the V and H subfamilies we did not find such an overrepresentation, likely due to lower gene numbers (V subfamily: Fisher’s test, odds ratio = 1.64, _p_ value = 0.17; H subfamily:
Fisher’s test, odds ratio = 1.71, _p_ value = 0.30). On the other hand, we found that 80% (51/64) of the OR genes overexpressed in nurses belong to the 9-exon subfamily. This results in an
overrepresentation of the 9-exon subfamily in genes that were overexpressed in nurses (Fisher’s test, odds ratio = 22.18, _p_ value < 0.001), with 45% (51/114) of this subfamily being
overexpressed in nurses. In contrast, only 6% (8/137) of the 9-exon subfamily was overexpressed in foragers, which is less than expected by chance (Fisher’s test, odds ratio = 0.09, _p_
value < 0.001). We also found that nurses overexpressed _Orco_ compared to foragers (FDR _p_ value = 0.001). GENES IN BIOGENIC AMINE PATHWAYS VARY IN EXPRESSION BETWEEN NURSES AND
FORAGERS Biogenic amines have been implicated in the regulation of behavior, and they may affect sensory perception46,87,88,89,90. We therefore screened our lists of brain and antennal DEGs
for biogenic amine pathway genes. We detected five differentially expressed genes in the antennae that are associated with biogenic amine signaling. Genes in the serotonin
(_5-hydroxytryptamine_, DBV15_11483), tyramine (_tyramine beta-hydroxylase_, DBV15_00422) and octopamine (_octopamine receptor_, LOC112465659) pathways were upregulated in the antennae of
foragers, while nurses showed a higher expression of genes in dopamine (_dopamine 1-like receptor 2_, DBV15_07611) and octopamine (_octopamine beta2 receptor_, DBV15_10418) pathways (Fig.
3A–E). These five genes were also expressed in the brain, but not differentially expressed between nurses and foragers. Also, no other biogenic amine pathway genes were found to be
differentially expressed in the brain. ASSOCIATION BETWEEN BEHAVIORAL VARIATION AND MOLECULAR PATHWAYS IN BRAIN AND ANTENNAE Our analysis revealed expression differences of genes expressed
in the brains of nurses and foragers involved in the regulation of task specialization in social insects. Previous studies have found that _Vg_ genes and the associated JH and IIS/TOR
pathways are important endocrine networks that play central roles in the regulation of lifespan, fertility and behavior in bees and ants36,91,92,93,94,95,96. Therefore, we searched and found
evidence in our RNA-seq data for task-associated expression of genes involved in the metabolism, biosynthesis, and regulation of these pathways in the brain and antennae (Table S1). These
genes included _conventional Vg_ (LOC112466671), _Vg-like A_ (named/classified as per;43 DBV15_03138), _venom carboxylesterase-6-like_ (DBV15_11528), and _protein takeout_ (DBV15_08771)
overexpressed in the brain of nurses. Meanwhile, _allatostatin A-like_ (LOC112454443) and _insulin-like growth factor I_ (_IGF1_) (LOC112454447) were found overexpressed in the brain of
foragers. Remarkably, _venom carboxylesterase-6-like_ and _IGF1_ were overexpressed in the antennae of nurses and foragers respectively. Additionally, high expression levels of the
zinc-finger transcription factor _Krüppel homolog-1_ (_Kr-h1_) were detected in the antennae of foragers (DBV15_06330) (Fig. 3F). The expression of this gene has been correlated with caste
and behavioral differences in the brain of social insects93,97,98,99,100, but little is known about its function and gene targets in the antennae. We performed a GO enrichment analysis to
gain a deeper understanding of the biological processes represented in the lists of DEGs. This analysis detected many enrichments based on single one or few genes, and we mention below a few
interesting processes with the number of genes driving the enrichment in parentheses (see Table S2 for complete list). Genes that were upregulated in the brain of nurses were enriched for
biological processes such as _translation_ (19 genes), _cellular iron ion homeostasis_ (2 genes), and _catabolic process_ (6 genes); while _translation_ (22 genes), _endocytosis_ (6 genes),
_glucose metabolic process_ (4 genes), and _regulation of cell cycle_ (3 genes) were enriched in the antennae. In the list of genes that were overexpressed in the brain of foragers, we found
that enriched processes included _peptide metabolic process_ (4 genes), _methionyl-tRNA aminoacylation_ (1 gene), _positive regulation of type I interferon production_ (1 gene), and _cell
redox homeostasis_ (1 gene); while _phosphatidylinositol phosphorylation_ (4 genes), _inositol phosphate dephosphorylation_ (3 genes), _carbohydrate metabolic process_ (17 genes), _innate
immune response_ (2 genes), and _nucleotide catabolic process_ (3 genes) were enriched in the antennae. DISCUSSION In this study, we found evidence supporting the hypothesis that variation
among social insect workers in their ability to perceive different chemical signals could contribute to the regulation of task allocation, and thus to DOL in insect societies. To do so, we
analyzed brain and antenna transcriptomes of nurses and foragers of the ant _T. longispinosus_. We report several lines of evidence that support our hypothesis. First, we found almost seven
times as many genes to be differentially expressed between nurses and foragers in the antennae as in the brain, indicating that peripheral sensory organs may have an important function in
the task specialization process of social insect workers. Second, we found that half of all OR genes of the _T. longispinosus_ genomes are differentially expressed between the antennae of
nurses and foragers, suggesting that behavioral specialization is associated with different sensory filters that result in specific perceptions of the chemical environment. Third, our
analyses revealed that nurses and foragers upregulated distinct families of OR genes, indicating that their sensory filters may target different types of chemical cues, possibly adjusted to
the tasks they perform. Finally, we detected several genes in multiple biogenic pathways to be differentially expressed between the antennae of nurses and foragers, potentially involved in
fine-tuning the sensitivity of the odorant filters. Many organisms have evolved sensory filters to focus on only a subset of all environmental cues101,102,103,104. In insects, the peripheral
olfactory filtering system enables individuals to detect and discriminate odors that convey ecologically relevant information used to mediate important behaviors such as courtship,
locomotion and navigation to avoid predators and locate food or nesting sites105. For example, the mosquito _Anopheles gambiae_ strongly responds to odorant components of its vertebrate
hosts that provide meals106. Sensory filters have been selected for because they limit the amount of information perceived by the organism, and thus the energy and time required by the brain
to process it105. Within insect colonies, individuals typically exhibit many morphological and physiological traits associated with increased efficiency in task specialization. These can be
understood as adaptations that allow individuals to become more proficient at their task due to learning, training, or the perception of valuable task-related information, allowing the
colony to avoid the cost of task switching107. In this context, it is interesting to find that _T. longispinosus_ workers may have different sensory filters that would serve as a basis for
their task specialization, as workers would mostly perceive chemical cues that pertain to their tasks. Our findings indicate that the sensory filter of ant workers is dynamic, and its
changes may underlie their behavioral maturation. In this study, we report a higher number of DEGs between nurses and foragers in the antennae than in the brain. The brain is a heterogeneous
tissue in which different cell types perform very specific functions, which differ greatly in their gene expression108,109,110. Therefore, using the whole brain for transcriptome analyses
could make it more difficult to identify genes that are differentially expressed only in specialized parts of the brains of nurses or foragers. In comparison, the antennae might have a more
uniform cell composition, which could facilitate the identification of DEGs. In contrast to this prediction, a transcriptome comparison of two different behavioral phenotypes of worker honey
bees revealed that transcriptional differences are much more pronounced in the antenna than in the separately studied brain parts including the mushroom body, antennal lobe and central
brain111. Furthermore, Chandra et al.112. used whole-brain RNA-seq in several ant species to detect differential expression of the gene _Ilp2_ between castes, a gene that was later found to
be expressed in only about 15 cells of the pars intercerebralis. This led us to conclude that it may be slightly more difficult to identify DEGs in the brain than in the antennae, but this
unlikely explains the nearly seven-fold difference in the number of DEGs. Age, genetic background, social environment, individual experiences and hormones influence behavioral differences
between workers leading to DOL13,20,23,113,114,115. In our study, we expect that age differed between individuals identified as nurses and those identified as foragers, although we did not
measure it directly. However, we know from previous studies on _T. longispinosus_ that workers live between one to three years and switch from brood carer to forager about one year after
eclosion, when new generation of workers emerged and taken over the care of the brood43,116. Previous experiments designed to disentangle gene expression associated with behavioral
specialization, age and fertility showed that behavioral specialization is much more strongly associated with gene expression than age and fertility in _T. longispinosus_39. We propose that
the molecular and physiological regulators such as JH, Vg, biogenic amines, and nutritional status known to regulate task specialization could drive different OR expression patterns, which
in turn would produce behavioral variation via contrasting abilities to detect different sets of odors. This hypothesis is supported by several studies (reviewed in ref. 117) showing that
the physiological condition of an animal can influence the level of receptor expression, including mating status, oviposition, feeding, circadian rhythm, experience, and aging.
Alternatively, we cannot exclude that the exposure to different odors that is associated with performing different tasks may at least in part have affected OR gene expression in the
antennae. However, such an effect is unlikely to explain the large-scale variation in gene expression, as there is very limited evidence that the mere exposure to odors influences the
expression of the gene coding for the OR that binds this odor118,119. According to our sensory filter hypothesis, ant workers would be expected to primarily detect chemical cues that
correspond to their tasks. We found that the 9-exon subfamily of OR was overrepresented in genes that were upregulated in the antennae of nurses. Interestingly, recent studies have shown a
rapid expansion of the 9-exon subfamily in ants, and several lines of evidence indicate that OR genes from this subfamily mediate complex social interactions in ant colonies67,68,69,70.
First, comparisons of antennal transcriptomes revealed that 9-exon OR genes are expressed more frequently in workers than in males, suggesting a role in social communication among
workers63,70. Second, representative OR from the 9-exon subfamily can detect CHC extracts from several castes71. According to McKenzie et al.70, 9-exon OR genes were first expressed in
solitary ancestors of aculeate wasps and facilitated CHC discrimination, likely for prey or mate recognition, with a lineage giving rise to the ancestors of ants. Moreover, OR genes of the
9-exon subfamily were convergently lost in socially parasitic ant species that lost the ability to perform brood care or foraging72, suggesting that they are essential for the performance of
these worker tasks. Our finding of an overexpression of 9-exon OR genes in the antennae of nurses is in line with these previous studies, and suggests that many of these receptors have
important functions within the nest, such as sensing chemical cues from the larvae, queen, or other workers, and/or that they are less important for sensing task-related stimuli or other
signals outside the nest. Among the OR genes overexpressed in nurses was also the co-receptor _Orco_, which is widely expressed in olfactory sensory neurons and nearly unchanged in sequence
in distant insect taxa120,121,122. ORs form a unique class of heteromeric cation channels composed of two related heptahelical subunits: a divergent OR subunit that confers odor specificity,
and the co-receptor _Orco_ subunit123,124. Since those functional receptors would increase the sensitivity of the workers to odors, we propose that the overexpression of _Orco_ may indicate
higher olfactory sensitivity to odors in the antennae of nurses compared to foragers. This would be supported by previous studies that showed that changes in _Orco_ expression can be
indicative of physiological conditions and sensory receptivity125,126. The behavioral transition from nursing to foraging may be triggered by a lower efficiency in detecting brood cues via
the downregulation of specific OR genes (e.g., from the 9-exon subfamily). This would result in ants moving farther away from the brood, and this change in spatial location may trigger the
behavioral transition to outside tasks127,128. In addition to being less efficient at detecting brood cues, the sensory filter of foragers may also become fine-tuned to detect a more diverse
set of odors. Foragers overexpressed a greater number of OR gene subfamilies compared to nurses (19 and 3 for foragers and nurses, respectively), which may indicate that the olfactory
system of foragers could perceive the more diverse chemical environment outside the nest. Similar to the 9-exon subfamily, the L subfamily has also been expanded in social insects64,66,67,
and along with the P and H subfamilies, it has been lost in socially parasitic ants72,129. Interestingly, the OR genes from the L, P, V, and H subfamilies have been upregulated in foragers,
and thus may have a task-specific function, such as recognition of chemical cues related to environmental perception or recruitment cues outside the nest. OR genes belonging to subfamilies
L, H and V have been shown to be highly responsive to long-chain hydrocarbons and are overexpressed in the antennae of males and workers of the ant _Harpegnathos saltator_130. In addition,
several ORs of the H subfamily have been proposed to act as putative floral odorant detectors in the antennae of honey bees131. Finding task-specific variation in OR gene expression raises
the question as to which molecular mechanisms regulate those changes. Variation in biogenic amines levels have been identified as one of the leading causes of behavioral plasticity and
specialization of social insects to different tasks (reviewed in ref. 132). Functional manipulation of biogenic amines has led to changes in behavior, dominance status and reproductive
activity, as well as shifts in worker task performance133,134,135,136. Previous studies on insects revealed that olfaction-guided behavior is mediated by biogenic amine receptors in the
antenna, and their expression is involved in fine-tuning the sensitivity of the olfactory system89,117. Signal transduction of biogenic amine receptors is mediated by G protein-coupled
receptors (GPCRs) located on the cell membrane, which trigger different signaling cascades that lead to increased or decreased cAMP level and Ca2+ release137,138,139,140. For example,
modified concentration cAMP and intracellular Ca2+ levels due to octopamine-induced signal transduction in the moth _Manduca sexta_141 activate _Orco_, leading to changes in ORN
sensitivity142,143. Our results suggest that the biogenic amine signaling pathway may modulate the sensory filtering function of insect antennae and alter sensitivity to various signals. We
found that genes encoding tyramine and its precursor, octopamine, are upregulated in forager antennae, similar to genes involved in serotonin signaling. Tyramine and dopamine (which was
upregulated in nurses) have been implicated in modulating taste and olfactory receptor neurons, while serotonin may serve as a neurotransmitter and neurohormone in antennal vessels and
mechanosensory organs89. Serotonin influences foraging activity88 and regulates food intake in many animals144,145,146. Dopamine signaling also plays an important role in controlling the
insect circadian clock and mediating clock-controlled behavioral phenotypes such as locomotion147,148. In our focal species _T. longispinosus_, inside workers were found to exhibit a
stronger circadian rhythmicity than foragers, which may be regulated via differences in the acetylation of histone proteins149. Changes in behavior and olfactory sensitivity in insects could
be related to the expression of genes involved in IIS, target of rapamycin (TOR), JH and Vg pathways, according to age, circadian rhythm, mating and feeding status117. For example, appetite
state in _D. melanogaster_ is signaled by insulin, which upregulates a peptide receptor on the olfactory receptor cells that innervate the DM1 glomerulus. Activation of the DM1 glomerulus
is enough to drive the fly to reach for food150. Recent studies have shown that pheromone release and odor sensitivity appear to be under JH control in _Schistocerca gregaria_ and _Locusta
migratoria_, which could lead to behavioral changes151,152,153,154,155. Finally, an experimental downregulation of _Vg-like A_ in _T. longispinosus_ workers resulted in decreased brood care
behavior and a lower sensitivity to brood-related chemical cues, suggesting changes in odor perception and olfactory-driven decision making43. Our results reveal that genes associated with
all these pathways were differentially expressed in the brain and antennae between nurses and foragers, predicting a link between their role in task-associated behavioral changes and the
regulation of odor perception. Given the central role of IIS, Vg, JH, and TOR pathways in regulating division of labor in social insects, and our finding of task-associated patterns of the
antennal expression of genes from multiple biogenic amines, we hypothesize that these modulators and hormones could be involved in the regulation of the olfactory filter. How the detailed
molecular mechanisms of these pathways in the brain are causally linked to the complex changes in olfactory perception in the antennae should therefore be investigated next. CONCLUSION Our
transcriptomic analyses of the brain and antennae of _T. longispinosus_ nurses and foragers provide support to our hypothesis that behavioral variation and task specialization in ant workers
are regulated via differences in olfactory perception. We predict that antennal physiology acts as sensory filters that limit the type and amount of chemical information passed to the
brain. This would allow workers to target relevant chemical information from the environment and discriminate signal from noise without using energetically costly processing by the central
nervous system. We argue that this sensory filter is flexible and regulated through changes in physiological conditions such as age, nutrition, and hormones. Variation among workers in their
efficiency to detect specific chemical cues would result in task specialization and division of labor. The information perceived by the peripheral ORs is transmitted to the primary brain
center of the olfactory pathway, the glomeruli of the antennal lobes. The question now arises whether there are differences between nurses and foragers in the morphology or physiology of the
antennal lobes. A limited subset of active ORs and glomeruli might be easier to process and less energy consuming. Our study opens novel avenues of research to better understand the role of
sensory filters in controlling DOL in insect societies. METHODS SAMPLE COLLECTION AND BEHAVIORAL DETERMINATION A total of seven colonies of the ant _T. longispinosus_ were selected with an
average colony size of 110 ± 31.5 workers (mean ± sd, Dataset S4) at the moment when the workers were sampled. The ants were collected in the forests of the Edmund Niles Huyck Preserve,
Renssellearville, NY, USA (42°31′41.0′′N 74°09′38.8′′W), in June of 2018 with permission. Upon collection, we housed each colony in a plaster-floored nesting box (43 cm × 28 cm × 10 cm)
divided into three chambers containing a single slide nest, in which the colony relocated. A slide nest is an artificial nesting site comprised of a small Plexiglas cavity sandwiched between
two glass microscope slides. Colonies were established at the Johannes Gutenberg University in Mainz, Germany, under a 14 h:10 h light:dark photoperiod at 18 °C to a 22 °C temperature. We
provided honey and water ad libitum and fed crickets to the colony twice a week. To allow for visible behavioral division of labor between workers of the two behavioral phenotypes, we
marked, observed and recaptured ants from inside and outside the nest. We defined foragers as workers that perform outside-nest tasks, including gathering and searching for food and water
and exploring the environment surrounding the nest, while nurses remained inside the dark nest and cared for the ant brood. A total of 69 workers inside (from the brood pile) and 76 workers
outside the nest were marked with fine colored metal wires (0.02 mm Elektrisola, Eckenhagen, Germany). To immobilize the workers, they were placed with their heads and part of the thorax in
a notch of a soft sponge without prior anesthesia. Then we marked the ants with a very thin loop around the petiole. It was then checked that the wires did not interfere with the ants’
movement. We performed behavioral observations every 2 h, four times a day for 5 days (total = 20 scans), in which we noted down how many times an individual performed brood care and
foraging behavior and the position in the nest (Table S3). Based on these behavioral observations, the marked individuals found outside the nest, exploring the surroundings for food or
water, were identified as foragers. These workers usually do not care for the brood and do not frequently reside on brood piles, as ant colonies organize themselves spatially in a way that
reduces contact between foragers and brood156. We identified nurses as workers that remain inside the nest in direct contact with brood and were unlikely to leave the nest. Foragers were
found in 42% ± 26% of the observations outside of the nest, whereas nurses spend only 1% ± 3% outside. In contrast, nurses were interacting with the brood in 54% ± 22% of the observations,
whereas we found that foragers only did this only in 2% ± 4% of the observations. We scanned the behavior of workers over 20 observations, albeit earlier studies have shown that a single
observation makes it possible to group _T. longispinosus_ and other ants reliably into nurses and foragers that differ in behavior116,156, gene expression39 and CHC composition43.
Furthermore, spatial location can alone can predict behavior in _Temnothorax_ workers157. We focused in this study on individuals highly specialized on either foraging or brood care. Workers
that performed both tasks regularly were not included in this study. After all observations were completed, the marked nurses and foragers were collected, directly frozen in liquid nitrogen
and stored at -80 °C until further processing for dissection and pooling according to behavioral state and colony. RNA EXTRACTION AND SEQUENCING For RNA extraction, we removed both antennae
and stored them in a 1.5 ml Eppendorf tube containing 50 μl TRIzol (Invitrogen), cut the head off and fixed it on a slide with melted dental wax. We then made an incision around the head
with a surgical scalpel and removed the head capsule with forceps to expose the intact brain. Finally, we carefully pulled the brain out of the head capsule and removed the remains of other
tissues that were connected to it. The dissected brain was transferred to a 1.5 ml Eppendorf tube containing 20 μl PBS. Each dissection was completed in less than 5 min to prevent RNA
degradation. We dissected brain and antennae tissues from 48 nurses and 49 foragers. We pooled the brains and antennae from seven workers from each behavioral state and colony. The only
exception was the “GO” colony (NY18 E110), for which we pooled only 6 brains and 12 antennae from 6 nurses (Dataset S4) due to the loss of one sample during the dissection process.
Immediately after dissection of each brain and antennae, the Eppendorf tubes were kept on dry ice while we dissected the remaining individuals. Brain and antennae tissues were homogenized
with a pestle. Sample brains were transferred separately to a 1.5 ml Eppendorf tube containing 50 μl of TRIzol. We added 50 μl chloroform to each brain and antenna samples, gently inverted
for 30 s and then centrifuged samples at 12,000 × _g_ for 15 min at 4 °C. We collected the resulting supernatant and precipitated RNA with 25 μl 70% ethanol. We conducted the subsequent RNA
extraction with the RNeasy Mini Kit (Qiagen), following the manufacturer’s instruction. The resulting 28 samples (14 brains and 14 antennae) were stored at −80 °C until library preparation.
RNA-seq libraries were prepared by Novogene Company Limited, Cambridge, UK, using the NEBNext Ultra RNA Library Prep Kit for Illumina according to the manufacturer’s protocol. After
amplification and purification, 28 libraries were sequenced on an Illumina NovaSeq 6000 S4 flow cell platform using a paired-end 150 bp. Approximately 43 million raw reads were generated
from each library (Dataset S4). GENE EXPRESSION ANALYSES Raw data obtained from Novogene were checked using FastQC v.0.11.9158, and Illumina adapters were removed using Trimmomatic
v.0.36159. The protein-coding genes of _T. longispinosus_ together with the manual OR annotations (GCA_004794745.1)160; and the congener _T. curvispinosus_ (GCA_003070985.1) were retrieved
from the NCBI database and we used Liftoff v.1.6.1 tool161 to assign these annotations to the recently published _T. longispinosus_ genome72. In total, 10,029 of 13,061 (~77%) annotated
protein-coding genes were assigned from the original _T. longispinosus_ assembly (genes identified as “DBV15”) and 4808 were assigned from _T. curvispinosus_ (genes identified as “LOC”), for
a total of 14,837. For gene expression analysis, reads were mapped to our _T. longispinosus_ genome assembly, and the read counts table was generated using STAR 2.7.0162 with default
settings. Detailed mapping statistics for each sample is available in Dataset S4. We used the deseq2 v1.16.1 package for R to identify differentially expressed genes163. To avoid biased
results due to low read counts, we removed from the counting matrix those genes for which less than 10 of the reads mapped to at least 6 of our 14 samples (_n_ − 1 of the smallest sample
size). Then, we conducted a differential gene expression analysis with DESeq2164. We began with comparisons between nurses and foragers using the ~Colony+Task model, followed by a likelihood
ratio test (LRT) approach, with colony ID as a fixed factor. Genes were considered differentially expressed if the false discovery rate (FDR), using Benjamini–Hochberg procedure, had an
adjusted _p_ value of ≤0.05. The resulting lists of DEGs refer to genes that are overexpressed and underexpressed in foragers compared to nurses. We used the online tool Venny v.2.1
(https://bioinfogp.cnb.csic.es/tools/venny) to generate a Venn diagram containing the DEGs associated with task and tissues. Separation of differentially expressed genes by task was
visualized by performing principal component analysis (PCA) with a 95% confidence ellipse using the ggplot2 v3.4.2 package for R165. For PCA, we used the transformed reads of filtered
transcriptomes from all contigs using the plotPCA function provided by DESeq2. Samples “GO”, “BG” and “YY” showed a divergent expression pattern. To ensure that our results were not
influenced by these deviating samples, we re-run the DEseq2 analyses repeatedly removing sample after sample. This resulted show slight shifts in the number of DEGs in the antennae (2267
with all samples vs. 1960 without “GO”; 1861 without “BG”; and 1862 without “YY”) and in the brain (339 with all samples vs. 227 without “GO”; 272 without “BG”; and 346 without “YY”).
However, the main findings remained similar and a large number of differentially expressed ORs were always found in all analyses (209 with all samples vs. 195 without “GO”; 209 without “BG”;
and 179 without “YY”). Finally, OR genes that were upregulated in each behavioral phenotype were visualized in a volcano plot using ggplot2. All statistical tests and graphical
visualizations were performed in RStudio v.1.4.1106166. IDENTIFICATION OF BEHAVIOR CANDIDATE GENES AND ORS We used gene annotations based on a BlastX search of the _T. longispinosus_
transcriptome compared to a list of different invertebrate proteomes (i.e., _Acromyrmex echinatior_, _Apis mellifera_, _Camponotus floridanus_, _Drosophila melanogaster_, _Harpegnathos
saltator_, _Odontomachus brunneus_, _Temnothorax curvispinosus_) downloaded from the NCBI database with an _E_-value of 1e−5 and below. Clusters containing more than one sequence match per
species were reduced to a single specimen based on the highest blast score. We constructed orthogroups across all of the above species using OrthoFinder167, including amino acid sequences
from the _T. longispinosus_ proteome160, and retained orthogroups containing caste DEGs (Dataset S5) to again compare potential behavioral candidate genes previously identified as involved
in regulating the division of labor in social insects39,42,94,100,155,168,169,170,171. GO enrichment analysis was performed with TopGO v.2.44.0 for R using a Fisher’s exact test for the
different gene sets compared to the whole genome with the weight01 algorithm172. Only annotated GO terms with a _p_ value of ≤0.05 were considered significantly enriched. OR protein sets
were clustered across multiple ant species using OrthoFinder to derive orthologous groups and identify subfamilies for each OR in _T. longispinosus_. To associate orthogroups with previously
identified OR subfamilies, we used OR annotation in _Atta cephalotes_, _Acromyrmex echinatior_ from Engsontia et al.67, and _Camponotus floridanus_, _Harpegnathos saltator_, and _Solenopsis
invicta_ from Zhou et al.63,66. Missing subfamily information was labeled as “unassigned” (Dataset S3). STATISTICS AND REPRODUCIBILITY The experiments were performed in seven replicates.
Each sample contains the pooled RNA from either the antennae or brain of seven ant workers of the respective behavioral phenotype belonging to the same colony, with the colony representing
the level of replicates. Bar graphs show next to median and quartiles, the individual data points. Statistical analyses were performed in RStudio v.1.4.1106, bioinformatics analyses in Bash,
and scripts for both are available on Mendeley. Gene expression analyses were performed using DEseq2, and to exclude the influence of outliers, these were removed individually, and results
presented without them. REPORTING SUMMARY Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY Numerical
source data found in Supplementary Data S1 was used to create Fig. 1A and Data S2 for Figs. 2B, C and 3. Raw sequencing reads generated for this study have been deposited in NCBI under
BioProject PRJNA926589. Any remaining information can be obtained from the corresponding author upon request. CODE AVAILABILITY Data analysis and visualization for this study was done using
code written in R and Bash, which can be found on Mendeley Data (https://doi.org/10.17632/yyg46xmph6.3). REFERENCES * Maynard Smith, J. & Szathmáry, E. _The Major Evolutionary
Transitions in Evolution_ (Oxford University Press, 1995). * Smith, A. _An Inquiry into the Nature and Causes of the Wealth of Nations_ (University Chicago Press, 1776). * Veening, J. W. et
al. Transient heterogeneity in extracellular protease production by _Bacillus subtilis_. _Mol. Syst. Biol._ 4, 1–15 (2008). Google Scholar * Marlow, V. L. et al. The prevalence and origin
of exoproteaseproducing cells in the _Bacillus subtilis_ biofilm. _Microbiology_ 160, 56–66 (2014). CAS PubMed PubMed Central Google Scholar * van Gestel, J., Vlamakis, H. & Kolter,
R. From cell differentiation to cell collectives: bacillus subtilis uses division of labor to migrate. _PLoS Biol._ 13, 1–29 (2015). Google Scholar * West, S. A. & Cooper, G. A.
Division of labour in microorganisms: an evolutionary perspective. _Nat. Rev. Microbiol._ 14, 716–723 (2016). CAS PubMed Google Scholar * Oster, G. F. & Wilson, E. O. Caste and
ecology in the social insects. _Acta Biotheor._ 28, 234–235 (1979). Google Scholar * Wilson, E. O. The ergonomics of caste in the social insects. _Am. Nat._ 102, 41–66 (1968). Google
Scholar * Wilson, E. O. Behavioral discretization and the number of castes in an ant species. _Behav. Ecol. Sociobiol._ 1, 141–154 (1976). Google Scholar * Herbers, J. M. Social social
organization in _Leptothorax_ ants: within-and between-species patterns. _Psyche J. Entomol._ 90, 52489 (1983). Google Scholar * Pinter-Wollman, N., Hubler, J., Holley, J. A., Franks, N. R.
& Dornhaus, A. How is activity distributed among and within tasks in _Temnothorax ants_? _Behav. Ecol. Sociobiol._ 66, 1407–1420 (2012). Google Scholar * Seeley, T. D. Adaptive
significance of the age polyethism schedule in honeybee colonies. _Behav. Ecol. Sociobiol._ 11, 287–293 (1982). Google Scholar * Tripet, F. & Nonacs, P. Foraging for work and age-based
polyethism: the roles of age and previous experience on task choice in ants. _Ethology_ 110, 863–877 (2004). Google Scholar * Blanchard, G. B., Orledge, G. M., Reynolds, S. E. & Franks,
N. R. Division of labour and seasonality in the ant _Leptothorax albipennis_: worker corpulence and its influence on behaviour. _Anim. Behav._ 59, 723–738 (2000). CAS PubMed Google
Scholar * Toth, A. L. & Robinson, G. E. Worker nutrition and division of labour in honeybees. _Anim. Behav._ 69, 427–435 (2005). Google Scholar * Wetterer, J. K. The ecology and
evolution of worker size-distribution in leaf-cutting ants (Hymenoptera: Formicidae). _Sociobiology_ 34, 119–144 (1999). Google Scholar * Fewell, J. H. & Page, R. E. Genotypic variation
in foraging responses to environmental stimuli by honey bees, _Apis mellifera_. _Experientia_ 49, 1106–1112 (1993). Google Scholar * Graham, S., Myerscough, M. R., Jones, J. C. &
Oldroyd, B. P. Modelling the role of intracolonial genetic diversity on regulation of brood temperature in honey bee (_Apis mellifera_ L.) colonies. _Insectes Soc._ 53, 226–232 (2006).
Google Scholar * Gove, R., Hayworth, M., Chhetri, M. & Rueppell, O. Division of labour and social insect colony performance in relation to task and mating number under two alternative
response threshold models. _Insectes Soc._ 56, 319–331 (2009). Google Scholar * Ravary, F., Lecoutey, E., Kaminski, G., Châline, N. & Jaisson, P. Individual experience alone can
generate lasting division of labor in ants. _Curr. Biol._ 17, 1308–1312 (2007). CAS PubMed Google Scholar * Gautrais, J., Theraulaz, G., Deneubourg, J. L. & Anderson, C. Emergent
polyethism as a consequence of increased colony size in insect societies. _J. Theor. Biol._ 215, 363–373 (2002). PubMed Google Scholar * Merkle, D. & Middendorf, M. Dynamic polyethism
and competition for tasks in threshold reinforcement models of social insects. _Adapt Behav._ 12, 251–262 (2004). Google Scholar * Jeanson, R., Fewell, J. H., Gorelick, R. & Bertram, S.
M. Emergence of increased division of labor as a function of group size. _Behav. Ecol. Sociobiol._ 62, 289–298 (2007). Google Scholar * Wilson, E. O. Caste and division of labor in
leaf-cutter ants (Hymenoptera: Formicidae: Atta). _Behav. Ecol. Sociobiol._ 7, 157–165 (1980). Google Scholar * Wakano, J. Y., Nakata, K. & Yamamura, N. Dynamic model of optimal age
polyethism in social insects under stable and fluctuating environments. _J. Theor. Biol._ 193, 153–165 (1998). Google Scholar * Iwasa, Y. & Yamaguchi, S. Task allocation in a
cooperative society: specialized castes or age-dependent switching among ant workers. _Sci. Rep._ 10, 1–9 (2020). Google Scholar * Robinson, E. J. H., Feinerman, O. & Franks, N. R.
Flexible task allocation and the organization of work in ants. _Proc. R. Soc. B Biol. Sci._ 276, 4373–4380 (2009). Google Scholar * Shimoji, H., Kasutani, N., Ogawa, S. & Hojo, M. K.
Worker propensity affects flexible task reversion in an ant. _Behav. Ecol. Sociobiol_. 74, 92 (2020). * Ben-Shahar, Y., Robichon, A., Sokolowski, M. B. & Robinson, G. E. Influence of
gene action across different time scales on behavior. _Science_ 296, 741–744 (2002). CAS PubMed Google Scholar * Ben-Shahar, Y., Leung, H. T., Pak, W. L., Sokolowski, M. B. &
Robinson, G. E. cGMP-dependent changes in phototaxis: a possible role for the foraging gene in honey bee division of labor. _J. Exp. Biol._ 206, 2507–2515 (2003). CAS PubMed Google Scholar
* Whitfield, C. W., Cziko, A. M. & Robinson, G. E. Gene expression profiles in the brain predict behavior in individual honey bees. _Science_ 302, 296–299 (2003). CAS PubMed Google
Scholar * Ingram, K. K., Oefner, P. & Gordon, D. M. Task-specific expression of the foraging gene in harvester ants. _Mol. Ecol._ 14, 813–818 (2005). CAS PubMed Google Scholar *
Lucas, C. & Sokolowski, M. B. Molecular basis for changes in behavioral state in ant social behaviors. _Proc. Natl Acad. Sci. USA_ 106, 6351–6356 (2009). CAS PubMed PubMed Central
Google Scholar * Zayed, A. & Robinson, G. E. Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. _Annu. Rev. Genet._ 46,
591–615 (2012). CAS PubMed Google Scholar * Sullivan, J. P., Jassim, O., Fahrbach, S. E. & Robinson, G. E. Juvenile hormone paces behavioral development in the adult worker honey bee.
_Horm. Behav._ 37, 1–14 (2000). CAS PubMed Google Scholar * Ament, S. A., Corona, M., Pollock, H. S. & Robinson, G. E. Insulin signaling is involved in the regulation of worker
division of labor in honey bee colonies. _Proc. Natl Acad. Sci. USA_ 105, 4226–4231 (2008). CAS PubMed PubMed Central Google Scholar * Dolezal, A. G., Brent, C. S., Hölldobler, B. &
Amdam, G. V. Worker division of labor and endocrine physiology are associated in the harvester ant, _Pogonomyrmex californicus_. _J. Exp. Biol._ 215, 454–460 (2012). CAS PubMed PubMed
Central Google Scholar * Corona, M. et al. Vitellogenin underwent subfunctionalization to acquire caste and behavioral specific expression in the harvester ant _Pogonomyrmex barbatus_.
_PLoS Genet_. 9, e1003730 (2013). * Kohlmeier, P., Alleman, A. R., Libbrecht, R., Foitzik, S. & Feldmeyer, B. Gene expression is more strongly associated with behavioural specialization
than with age or fertility in ant workers. _Mol. Ecol._ 28, 658–670 (2019). PubMed Google Scholar * Libbrecht, R. et al. Interplay between insulin signaling, juvenile hormone, and
vitellogenin regulates maternal effects on polyphenism in ants. _Proc. Natl Acad. Sci. USA_ 110, 11050–11055 (2013). CAS PubMed PubMed Central Google Scholar * Marco Antonio, D. S.,
Guidugli-Lazzarini, K. R., Do Nascimento, A. M., Simões, Z. L. P. & Hartfelder, K. RNAi-mediated silencing of vitellogenin gene function turns honeybee (_Apis mellifera_) workers into
extremely precocious foragers. _Naturwissenschaften_ 95, 953–961 (2008). CAS PubMed Google Scholar * Gospocic, J. et al. The neuropeptide corazonin controls social behavior and caste
Identity in ants. _Cell_ 170, 748.e12–759.e12 (2017). Google Scholar * Kohlmeier, P., Feldmeyer, B. & Foitzik, S. _Vitellogenin-like_ A–associated shifts in social cue responsiveness
regulate behavioral task specialization in an ant. _PLoS Biol._ 16, 1–26 (2018). Google Scholar * Kamhi, J. F. & Traniello, J. F. A. Biogenic amines and collective organization in a
superorganism: neuromodulation of social behavior in ants. _Brain Behav. Evol._ 82, 220–236 (2013). PubMed Google Scholar * Schulz, D. & Robinson, G. Biogenic amines and division of
labor in honey bee colonies: behaviorally related changes in the antennal lobes and age-related changes in the mushroom bodies. _J. Comp. Physiol. A_ 184, 481–488 (1999). * Mercer, A. R.
& Menzel, R. The effects of biogenic amines on conditioned and unconditioned responses to olfactory stimuli in the honeybee _Apis mellifera_. _J. Comp. Physiol._ 145, 363–368 (1982). CAS
Google Scholar * Scheiner, R., Entler, B. V., Barron, A. B., Scholl, C. & Thamm, M. The effects of fat body tyramine level on gustatory responsiveness of honeybees (_Apis mellifera_)
differ between behavioral castes. _Front. Syst. Neurosci._ 11, 1–8 (2017). Google Scholar * Ferguson, S. T., Bakis, I. & Zwiebel, L. J. Advances in the study of olfaction in eusocial
ants. _Insects_ 12, 252 (2021). PubMed PubMed Central Google Scholar * Martin, S. & Drijfhout, F. A review of ant cuticular hydrocarbons. _J. Chem. Ecol._ 35, 1151–1161 (2009). CAS
PubMed Google Scholar * van Wilgenburg, E., Symonds, M. R. E. & Elgar, M. A. Evolution of cuticular hydrocarbon diversity in ants. _J. Evol. Biol._ 24, 1188–1198 (2011). PubMed Google
Scholar * Slifer, E. H. The structure of arthropod chemoreceptors. _Annu. Rev. Entomol._ 15, 121–142 (1970). Google Scholar * Altner, H. & Prillinger, L. Ultrastructure of
invertebrate chemo-, thermo-, and hygroreceptors and its functional significance. _Int. Rev. Cytol._ 67, 69–139 (1980). Google Scholar * Hallberg, E. & Hansson, B. S. Arthropod
sensilla: morphology and phylogenetic considerations. _Microsc. Res. Tech._ 47, 428–439 (1999). CAS PubMed Google Scholar * Wicher, D. & Miazzi, F. Functional properties of insect
olfactory receptors: ionotropic receptors and odorant receptors. _Cell Tissue Res._ 383, 7–19 (2021). CAS PubMed PubMed Central Google Scholar * Leal, W. S. Odorant reception in insects:
roles of receptors, binding proteins, and degrading enzymes. _Annu. Rev. Entomol._ 58, 373–391 (2013). CAS PubMed Google Scholar * Sánchez‐Gracia, A., Vieira, F. G., Almeida, F. C. &
Rozas, J. Comparative genomics of the major chemosensory gene families in arthropods. _Encyclopedia Life Sci_. https://doi.org/10.1002/9780470015902.a0022848 (2011). * Breer, H., Fleischer,
J., Pregitzer, P. & Krieger, J. in _Olfactory Concepts of insect Control-Alternative to Insecticides_ (ed. Picimbon, J.-F.) 93–114 (Springer, 2019). * Jacquin-Joly, E. & Lucas, P.
Pheromone reception and transduction: mammals and insects illustrate converging mechanisms across phyla. _Curr. Top. Neurochem._ 4, 75–105 (2005). CAS Google Scholar * Wilson, R. K. &
Mainen, Z. F. Early events in olfactory processing. _Annu. Rev. Neurosci._ 29, 163–201 (2006). CAS PubMed Google Scholar * Anton, S. & Homberg, U. in _Insect Olfaction_ (ed. Hansson,
B.S.) 97–124 (Springer, 1999). * Hansson, B. S. & Anton, S. Function and morphology of the antennal lobe: new developments. _Annu. Rev. Entomol._ 45, 203–231 (2000). CAS PubMed Google
Scholar * de Belle, J. S. & Kanzaki, R. in _Insect Olfaction_ (ed. Hansson, B.S.) 243–281 (Springer, 1999). * Zhou, X. et al. Phylogenetic and transcriptomic analysis of chemosensory
receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. _PLoS Genet_. 8, e1002930 (2012). * Saad, R., Cohanim, A. B., Kosloff, M. & Privman, E.
Neofunctionalization in ligand binding sites of ant olfactory receptors. _Genome Biol. Evol._ 10, 2490–2500 (2018). CAS PubMed PubMed Central Google Scholar * Oxley, P. R. et al. The
genome of the clonal raider ant _Cerapachys Biroi_. _Curr. Biol._ 24, 451–458 (2014). CAS PubMed PubMed Central Google Scholar * Zhou, X. et al. Chemoreceptor evolution in Hymenoptera
and its implications for the evolution of eusociality. _Genome Biol. Evol._ 7, 2407–2416 (2015). CAS PubMed PubMed Central Google Scholar * Engsontia, P., Sangket, U., Robertson, H. M.
& Satasook, C. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. _BMC Res. Notes_ 8, 1–13 (2015). Google
Scholar * Smith, C. D. et al. Draft genome of the globally widespread and invasive Argentine ant (_Linepithema humile_). _Proc. Natl Acad. Sci. USA_ 108, 5673–5678 (2011). CAS PubMed
PubMed Central Google Scholar * Smith, C. R. et al. Draft genome of the red harvester ant _Pogonomyrmex barbatus_. _Proc. Natl Acad. Sci. USA_ 108, 5667–5672 (2011). CAS PubMed PubMed
Central Google Scholar * McKenzie, S. K., Fetter-Pruneda, I., Ruta, V. & Kronauer, D. J. C. Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of
odorant receptors in chemical communication. _Proc. Natl Acad. Sci. USA_ 113, 14091–14096 (2016). CAS PubMed PubMed Central Google Scholar * Pask, G. M. et al. Specialized odorant
receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. _Nat. Commun._ 8, 1–10 (2017). CAS Google Scholar * Jongepier, E. et al. Convergent loss of
chemoreceptors across independent origins of slave-making in ants. _Mol. Biol. Evol._ 39, 1–14 (2022). Google Scholar * Trible, W. et al. _orco_ Mutagenesis causes loss of antennal lobe
glomeruli and impaired social behavior in ants. _Cell_ 170, 727.e10–735.e10 (2017). Google Scholar * Yan, H. et al. An engineered _orco_ mutation produces aberrant social behavior and
defective neural development in ants. _Cell_ 170, 736.e9–747.e9 (2017). Google Scholar * Beshers, S. & Fewell, J. H. Models of division of labor in social insects. _Annu. Rev. Entomol._
46, 413–440 (2001). CAS PubMed Google Scholar * Lichocki, P., Tarapore, D., Keller, L. & Floreano, D. Neural networks as mechanisms to regulate division of labor. _Am. Nat._ 179,
391–400 (2012). PubMed Google Scholar * Naug, D. From division of labor to collective behavior: behavioral analyses at different levels. _Behav. Ecol. Sociobiol._ 70, 1113–1115 (2016).
Google Scholar * Jeanson, R. & Weidenmüller, A. Interindividual variability in social insects - proximate causes and ultimate consequences. _Biol. Rev._ 89, 671–687 (2014). PubMed
Google Scholar * Ulrich, Y. et al. Response thresholds alone cannot explain empirical patterns of division of labor in social insects. _PLoS Biol._ 19, 1–21 (2021). Google Scholar *
Yamanaka, O., Shiraishi, M., Awazu, A. & Nishimori, H. Verification of mathematical models of response threshold through statistical characterisation of the foraging activity in ant
societies. _Sci. Rep._ 9, 1–8 (2019). Google Scholar * Shiraishi, M., Yamanaka, O. & Nishimori, H. Effect of interaction network structure in a response threshold model. _Artif. Life
Robot._ 27, 743–750 (2022). Google Scholar * Theraulaz, G., Bonabeau, E. & Deneubourg, J. L. Response threshold reinforcement and division of labour in insect societies. _Proc. R. Soc.
B Biol. Sci._ 265, 327–332 (1998). Google Scholar * Bonabeau, E., Theraulaz, G. & Deneubourg, J. L. Quantitative study of the fixed threshold model for the regulation of division of
labour in insect societies. _Proc. R. Soc. B Biol. Sci._ 263, 1565–1569 (1996). Google Scholar * Bonabeau, E., Theraulaz, G. & Deneubourg, J. L. Fixed response thresholds and the
regulation of division of labor in insect societies. _Bull. Math. Biol._ 60, 753–807 (1998). Google Scholar * Bonabeau, E., Sobkowski, A., Theraulaz, G. & Deneubourg, J. L. Adaptive
task allocation inspired by a model of division of labor in social insects. In: _Proc. BCEC97_ 36–45 (ACM, 1997). * Thiagarajan, D. & Sachse, S. Multimodal information processing and
associative learning in the insect brain. _Insects_ 13, 332 (2022). PubMed PubMed Central Google Scholar * Raza, M. F. et al. Biogenic amines mediate learning success in appetitive odor
conditioning in honeybees. _J. King Saud. Univ. Sci._ 34, 0–6 (2022). CAS Google Scholar * Schulz, D. J., Elekonich, M. M. & Robinson, G. E. Biogenic amines in the antennal lobes and
the initiation and maintenance of foraging behavior in honey bees. _J. Neurobiol._ 54, 406–416 (2003). CAS PubMed Google Scholar * Zhukovskaya, M. I. & Polyanovsky, A. D. Biogenic
amines in insect antennae. _Front. Syst. Neurosci._ 11, 1–9 (2017). Google Scholar * Cook, C. N. et al. Individual differences in learning and biogenic amine levels influence the
behavioural division between foraging honeybee scouts and recruits. _J. Anim. Ecol._ 88, 236–246 (2019). PubMed Google Scholar * Azevedo, D. O., De Paula, S. O., Zanuncio, J. C., Martinez,
L. C. & Serrão, J. E. Juvenile hormone downregulates vitellogenin production in _Ectatomma tuberculatum_ (Hymenoptera: Formicidae) sterile workers. _J. Exp. Biol._ 219, 103–108 (2016).
PubMed Google Scholar * Rodrigues, M. A. & Flatt, T. Endocrine uncoupling of the trade-off between reproduction and somatic maintenance in eusocial insects. _Curr. Opin. Insect Sci._
16, 1–8 (2016). PubMed Google Scholar * Jedlicka, P., Ernst, U. R., Votavová, A., Hanus, R. & Valterová, I. Gene expression dynamics in major endocrine regulatory pathways along the
transition from solitary to social life in a bumblebee, _Bombus terrestris_. _Front. Physiol._ 7, 1–20 (2016). Google Scholar * Opachaloemphan, C. et al. Early behavioral and molecular
events leading to caste switching in the ant _Harpegnathos_. _Genes Dev._ 35, 410–424 (2021). CAS PubMed PubMed Central Google Scholar * Norman, V. C. & Hughes, W. O. H. Behavioural
effects of juvenile hormone and their influence on division of labour in leaf-cutting ant societies. _J. Exp. Biol._ 219, 8–11 (2016). PubMed Google Scholar * Robinson, G. E. Regulation of
honey bee age polyethism by juvenile hormone. _Behav. Ecol. Sociobiol._ 20, 329–338 (1987). Google Scholar * Fussnecker, B. & Grozinger, C. Dissecting the role of Kr-h1 brain gene
expression in foraging behavior in honey bees (_Apis mellifera_). _Insect Mol. Biol._ 17, 515–522 (2008). CAS PubMed Google Scholar * Grozinger, C. M. & Robinson, G. E. Endocrine
modulation of a pheromone-responsive gene in the honey bee brain. _J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol._ 193, 461–470 (2007). CAS PubMed Google Scholar * Shpigler,
H. et al. The transcription factor Krüppel homolog 1 is linked to hormone mediated social organization in bees. _BMC Evol. Biol._ 10, 120 (2010). PubMed PubMed Central Google Scholar *
Gospocic, J. et al. Kr-h1 maintains distinct caste-specific neurotranscriptomes in response to socially regulated hormones. _Cell_ 184, 5807–5823.e14 (2021). CAS PubMed PubMed Central
Google Scholar * Warrant, E. J. Sensory matched filters. _Curr. Biol._ 26, 976–980 (2016). Google Scholar * Ponnath, A., Ryan, M. J., Fang, Z. & Farris, H. E. Tuned in to communication
sounds: neuronal sensitivity in the túngara frog midbrain to frequency modulated signals. _PLoS ONE_ 17, e0268383 (2022). CAS PubMed PubMed Central Google Scholar * Hempel De Ibarra,
N., Holtze, S., Bäucker, C., Sprau, P. & Vorobyev, M. The role of colour patterns for the recognition of flowers by bees. _Philos. Trans. R. Soc. B Biol. Sci._ 377, 20210284 (2022).
Google Scholar * Patullo, B. W. & Macmillan, D. L. Making sense of electrical sense in crayfish. _J. Exp. Biol._ 213, 651–657 (2010). CAS PubMed Google Scholar * von der Emde, G.
& Warrant, E. _The Ecology of Animal Senses Matched Filters for Cconomical Sensing_ 1st edn (Springer International Publishing, 2016). * Carey, A. F., Wang, G., Su, C. Y., Zwiebel, L. J.
& Carlson, J. R. Odorant reception in the malaria mosquito _Anopheles gambiae_. _Nature_ 464, 66–71 (2010). CAS PubMed PubMed Central Google Scholar * Dornhaus, A. Specialization
does not predict individual efficiency in an ant. _PLoS Biol._ 6, 2368–2375 (2008). CAS Google Scholar * Koch, C. & Laurent, G. Complexity and the nervous system. _Science_ 284, 96–98
(1999). CAS PubMed Google Scholar * McCaffrey, J. B. The brain’s heterogeneous functional landscape. _Philos. Sci._ 82, 1010–1022 (2015). Google Scholar * Ito, M., Masuda, N., Shinomiya,
K., Endo, K. & Ito, K. Systematic analysis of neural projections reveals clonal composition of the _Drosophila_ brain. _Curr. Biol._ 23, 644–655 (2013). CAS PubMed Google Scholar *
Kennedy, A. et al. Use of waggle dance information in honey bees is linked to gene expression in the antennae, but not in the brain. _Mol. Ecol._ 30, 2676–2688 (2021). PubMed Google Scholar
* Chandra, V. et al. Social regulation of insulin signaling and the evolution of eusociality in ants. _Science_ 361, 398–402 (2018). CAS PubMed PubMed Central Google Scholar *
Libbrecht, R. & Keller, L. Genetic compatibility affects division of labor in the argentine ant _Linepithema humile_. _Evolution_ 67, 517–524 (2013). PubMed Google Scholar * Schulz, D.
J., Sullivan, J. P. & Robinson, G. E. Juvenile hormone and octopamine in the regulation of division of labor in honey bee colonies. _Horm. Behav._ 42, 222–231 (2002). CAS PubMed
Google Scholar * Geva, S., Hartfelder, K. & Bloch, G. Reproductive division of labor, dominance, and ecdysteroid levels in hemolymph and ovary of the bumble bee _Bombus terrestris_. _J.
Insect Physiol._ 51, 811–823 (2005). CAS PubMed Google Scholar * Kohlmeier, P. et al. Intrinsic worker mortality depends on behavioral caste and the queens’ presence in a social insect.
_Sci. Nat_. 104, 34 (2017). * Gadenne, C., Barrozo, R. B. & Anton, S. Plasticity in insect olfaction: to smell or not to smell? _Annu. Rev. Entomol._ 61, 317–333 (2016). CAS PubMed
Google Scholar * Zhou, S., Stone, E. A., Mackay, T. F. C. & Anholt, R. R. H. Plasticity of the chemoreceptor repertoire in _Drosophila melanogaster_. _PLoS Genet_. 5, e1000681 (2009). *
Von Der Weid, B. et al. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. _Nat. Neurosci._ 18, 1455–1463 (2015). PubMed Google Scholar
* Benton, R., Sachse, S., Michnick, S. W. & Vosshall, L. B. Atypical membrane topology and heteromeric function of _Drosophila_ odorant receptors in vivo. _PLoS Biol._ 4, 240–257
(2006). CAS Google Scholar * Krieger, J., Klink, O., Mohl, C., Raming, K. & Breer, H. A candidate olfactory receptor subtype highly conserved across different insect orders. _J. Comp.
Physiol. A Neuroethol. Sens. Neural Behav. Physiol._ 189, 519–526 (2003). CAS PubMed Google Scholar * Larsson, M. C. et al. Or83b encodes a broadly expressed odorant receptor essential
for _Drosophila_ olfaction. _Neuron_ 43, 703–714 (2004). CAS PubMed Google Scholar * Guo, H., Kunwar, K. & Smith, D. Odorant receptor sensitivity modulation in _Drosophila_. _J.
Neurosci._ 37, 9465–9473 (2017). CAS PubMed PubMed Central Google Scholar * Castillo, P., Husseneder, C. & Sun, Q. Molecular characterization and expression variation of the odorant
receptor co-receptor in the _Formosan subterranean_ termite. _PLoS ONE_ 17, e0267841 (2022). CAS PubMed PubMed Central Google Scholar * Sato, K. et al. Insect olfactory receptors are
heteromeric ligand-gated ion channels. _Nature_ 452, 1002–1006 (2008). CAS PubMed Google Scholar * del Mármol, J., Yedlin, M. A. & Ruta, V. The structural basis of odorant recognition
in insect olfactory receptors. _Nature_ 597, 126–131 (2021). PubMed PubMed Central Google Scholar * Mersch, D. P., Crespi, A. & Keller, L. Tracking individuals shows spatial fidelity
is a key regulator of ant social organization. _Science_ 340, 1090–1093 (2013). CAS PubMed Google Scholar * Richardson, T. O. et al. Ant behavioral maturation is mediated by a stochastic
transition between two fundamental states. _Curr. Biol._ 31, 2253–2260.e3 (2021). CAS PubMed Google Scholar * Schrader, L. et al. Relaxed selection underlies genome erosion in socially
parasitic ant species. _Nat. Commun._ 12, 1–13 (2021). Google Scholar * Slone, J. D. et al. Functional characterization of odorant receptors in the ponerine ant, _Harpegnathos saltator_.
_Proc. Natl Acad. Sci. USA_ 114, 8586–8591 (2017). CAS PubMed PubMed Central Google Scholar * Claudianos, C. et al. Odor memories regulate olfactory receptor expression in the sensory
periphery. _Eur. J. Neurosci._ 39, 1642–1654 (2014). PubMed Google Scholar * Barbero, F., Mannino, G. & Casacci, L. P. The role of biogenic amines in social insects: with a special
focus on ants. _Insects_ 14, 386 (2023). PubMed PubMed Central Google Scholar * Penick, C. A., Brent, C. S., Dolezal, K. & Liebig, J. Neurohormonal changes associated with ritualized
combat and the formation of a reproductive hierarchy in the ant _Harpegnathos saltator_. _J. Exp. Biol._ 217, 1496–1503 (2014). PubMed Google Scholar * Fussnecker, B. L., Smith, B. H.
& Mustard, J. A. Octopamine and tyramine influence the behavioral profile of locomotor activity in the honey bee (_Apis mellifera_). _J. Insect Physiol._ 52, 1083–1092 (2006). CAS
PubMed PubMed Central Google Scholar * Yakovlev, I. K. Effects of octopamine on aggressive behavior in red wood ants. _Neurosci. Behav. Physiol._ 48, 279–288 (2018). CAS Google Scholar
* Yaguchi, H., Inoue, T., Sasaki, K. & Maekawa, K. Dopamine regulates termite soldier differentiation through trophallactic behaviours. _R. Soc. Open Sci._ 3, 150574 (2016). PubMed
PubMed Central Google Scholar * Ohta, H. & Ozoe, Y. in _Advances in Insect Physiology_ 1st edn, Vol. 46 (ed, Cohen, E.) 73–166 (Elsevier Inc., 2014). * Beggs, K. T., Tyndall, J. D. A.
& Mercer, A. R. Honey bee dopamine and octopamine receptors linked to intracellular calcium signaling have a close phylogenetic and pharmacological relationship. _PLoS ONE_ 6, e26809
(2011). CAS PubMed PubMed Central Google Scholar * Blenau, W. & Baumann, A. Molecular and pharmacological properties of insect biogenic amine receptors: lessons from _Drosophila
melanogaster_ and _Apis mellifera_. _Arch. Insect Biochem. Physiol._ 48, 13–38 (2001). CAS PubMed Google Scholar * Vleugels, R., Verlinden, H. & Broeck, J. V. Serotonin, serotonin
receptors and their actions in insects. _Neurotransmitter_ 2, e314 (2015). Google Scholar * Dacks, A. M., Dacks, J. B., Christensen, T. A. & Nighorn, A. J. The cloning of one putative
octopamine receptor and two putative serotonin receptors from the tobacco hawkmoth, _Manduca sexta_. _Insect Biochem. Mol. Biol._ 36, 741–747 (2006). CAS PubMed PubMed Central Google
Scholar * Getahun, M. N., Olsson, S. B., Lavista-Llanos, S., Hansson, B. S. & Wicher, D. Insect odorant response sensitivity is tuned by metabotropically autoregulated olfactory
receptors. _PLoS ONE_ 8, 1–9 (2013). Google Scholar * Stengl, M. & Funk, N. W. The role of the coreceptor Orco in insect olfactory transduction. _J. Comp. Physiol. A Neuroethol. Sens.
Neural Behav. Physiol._ 199, 897–909 (2013). CAS PubMed Google Scholar * Blundell, J. E. & Halford, J. C. G. Serotonin and appetite regulation: implications for the pharmacological
treatment of obesity. _CNS Drugs_ 9, 473–495 (1998). CAS Google Scholar * French, A. S. et al. The role of serotonin in feeding and gut contractions in the honeybee. _J. Insect Physiol._
61, 8–15 (2014). CAS PubMed PubMed Central Google Scholar * Voigt, J. P. & Fink, H. Serotonin controlling feeding and satiety. _Behav. Brain Res._ 277, 14–31 (2015). CAS PubMed
Google Scholar * Beninger, R. J. The role of dopamine in locomotor activity and learning. _Brain Res. Rev._ 6, 173–196 (1983). CAS Google Scholar * Liang, X. et al. Morning and evening
circadian pacemakers independently drive premotor centers via a specific dopamine relay. _Neuron_ 102, 843.e4–857.e4 (2019). Google Scholar * Libbrecht, R., Nadrau, D. & Foitzik, S. A
role of histone acetylation in the regulation of circadian rhythm in ants. _iScience_ 23, 100846 (2020). PubMed PubMed Central Google Scholar * Root, C. M., Ko, K. I., Jafari, A. &
Wang, J. W. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. _Cell_ 145, 133–144 (2011). CAS PubMed PubMed Central Google Scholar * Anton, S., Dufour,
M. C. & Gadenne, C. P. Plasticity of olfactory-guided behaviour and its neurobiological basis: lessons from moths and locusts. _Entomol. Exp. Appl_. 123, 1–11 (2007). * Tawfik, A. I.,
Osir, E. O., Hassanali, A. & Ismail, S. H. Effects of juvenile hormone treatment on phase changes and pheromone production the desert locust, _Schistocerca gregaria_ (Forskal)
(Orthoptera: Acrididae). _J. Insect Physiol._ 43, 1177–1182 (1997). CAS PubMed Google Scholar * Tawfik, A. I., Treiblmayr, K., Hassanali, A. & Osir, E. O. Time-course haemolymph
juvenile hormone titres in solitarious and gregarious adults of _Schistocerca gregaria_, and their relation to pheromone emission, CA volumetric changes and oocyte growth. _J. Insect
Physiol._ 46, 1143–1150 (2000). CAS PubMed Google Scholar * Wiesel, G., Tappermann, S. & Dorn, A. Effects of juvenile hormone and juvenile hormone analogues on the phase behaviour of
_Schistocerca gregaria_ and _Locusta migratoria_. _J. Insect Physiol._ 42, 385–395 (1996). CAS Google Scholar * Guo, W. et al. Juvenile hormone suppresses aggregation behavior through
influencing antennal gene expression in locusts. _PLoS Genet._ 16, 1–18 (2020). Google Scholar * Pamminger, T., Foitzik, S., Kaufmann, K. C., Schützler, N. & Menzel, F. Worker
personality and its association with spatially structured division of labor. _PLoS ONE_ 9, 1–8 (2014). Google Scholar * Sendova-Franks, A. & Franks, N. R. Task allocation in ant
colonies within variable environments (a study of temporal polyethism: experimental). _Bull. Math. Biol._ 55, 75–96 (1993). Google Scholar * Andrews, S. A quality control tool for high
throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2016). * Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence
data. _Bioinformatics_ 30, 2114–2120 (2014). CAS PubMed PubMed Central Google Scholar * Kaur, R. et al. Ant behaviour and brain gene expression of defending hosts depend on the
ecological success of the intruding social parasite. _Philos. Trans. R. Soc. B Biol. Sci._ 374, 1769 (2019). Google Scholar * Shumate, A. & Salzberg, S. L. Liftoff: accurate mapping of
gene annotations. _Bioinformatics_ 37, 1639–1643 (2021). CAS PubMed PubMed Central Google Scholar * Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. _Bioinformatics_ 29, 15–21
(2013). CAS PubMed Google Scholar * Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. _Genome Biol._ 15, 550
(2014). PubMed PubMed Central Google Scholar * Love, M. I., Anders, S. & Huber, W. Differential analysis of count data - the DESeq2 package. _Genome Biol._ 15, 550 (2014). PubMed
PubMed Central Google Scholar * Wickham, H. ggplot2: Elegant graphics for data analysis. https://ggplot2.tidyverse.org (2016). * RStudio Team. RStudio: integrated development for R.
http://www.rstudio.com/ (2020). * Emms, D. M. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. _Genome
Biol._ 16, 157 (2015). PubMed PubMed Central Google Scholar * Das, B. & de Bekker, C. Time-course RNASeq of _Camponotus floridanus_ forager and nurse ant brains indicate links
between plasticity in the biological clock and behavioral division of labor. _BMC Genomics._ 23, 1–23 (2022). CAS Google Scholar * Alleman, A., Stoldt, M., Feldmeyer, B. & Foitzik, S.
Tandem-running and scouting behaviour are characterized by up-regulation of learning and memory formation genes within the ant brain. _Mol. Ecol._ 28, 2342–2359 (2019). PubMed Google
Scholar * Korb, J. et al. Comparative transcriptomic analysis of the mechanisms underpinning ageing and fecundity in social insects. _Philos. Trans. R. Soc. B Biol. Sci_. 376, 20190728
(2021). * Qiu, B. et al. Canalized gene expression during development mediates caste differentiation in ants. _Nat. Ecol. Evol._ 6, 1753–1765 (2022). PubMed PubMed Central Google Scholar
* Alexa, A. & Rahnenfuhrer, J. topGO: enrichment analysis for gene ontology. R package version 2.50.0. https://bioconductor.org/packages/release/bioc/html/topGO.html (2018). Download
references ACKNOWLEDGEMENTS We thank Diego Páez-Moscoso for many helpful discussions and comments throughout the study, Marah Stoldt and Carlotta Martelli for providing helpful comments on
the manuscript, Marion Kever and Jenny Fuchs for technical assistance, and Mark Harrison for providing the manual OR annotations. This project was funded by the Deutsche
Forschungsgemeinschaft (DFG, German Research Foundation) to SF – GRK2526/1 – Projectnr. 407023052. M.A.C. thanks the E.N. Huyck Preserve for support and funding. FUNDING Open Access funding
enabled and organized by Projekt DEAL. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Institute of Organismic and Molecular Evolution, Johannes Gutenberg University Mainz, Mainz, Germany
Marcel A. Caminer, Romain Libbrecht, Megha Majoe & Susanne Foitzik * Institut de Recherche sur la Biologie de l’Insecte, UMR 7261, CNRS, University of Tours, Tours, France Romain
Libbrecht * Institute of Developmental and Neurobiology, Johannes Gutenberg University Mainz, Mainz, Germany David V. Ho & Peter Baumann * Institute of Molecular Biology, Mainz, Germany
Peter Baumann Authors * Marcel A. Caminer View author publications You can also search for this author inPubMed Google Scholar * Romain Libbrecht View author publications You can also search
for this author inPubMed Google Scholar * Megha Majoe View author publications You can also search for this author inPubMed Google Scholar * David V. Ho View author publications You can
also search for this author inPubMed Google Scholar * Peter Baumann View author publications You can also search for this author inPubMed Google Scholar * Susanne Foitzik View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.A.C., R.L., and S.F. conceived the study; M.A.C. conducted the research; M.A.C., M.M., R.L., and
D.V.H. contributed to the development of the data analytical protocol; M.A.C. analyzed the data; and M.A.C., R.L., M.M., P.B., and S.F. wrote the manuscript. CORRESPONDING AUTHOR
Correspondence to Marcel A. Caminer. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Communications Biology_ thanks
Ching-Han Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editors: Luke Grinham, Karli Montague-Cardoso, David Favero.
ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION
SUPPORTING INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY DATA DATASET S1 DATASET S2 DATASET S3 DATASET S4 DATASET S5 REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article
is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give
appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in
this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative
Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a
copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Caminer, M.A., Libbrecht, R., Majoe, M. _et al._
Task-specific odorant receptor expression in worker antennae indicates that sensory filters regulate division of labor in ants. _Commun Biol_ 6, 1004 (2023).
https://doi.org/10.1038/s42003-023-05273-4 Download citation * Received: 18 April 2023 * Accepted: 22 August 2023 * Published: 02 October 2023 * DOI:
https://doi.org/10.1038/s42003-023-05273-4 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
