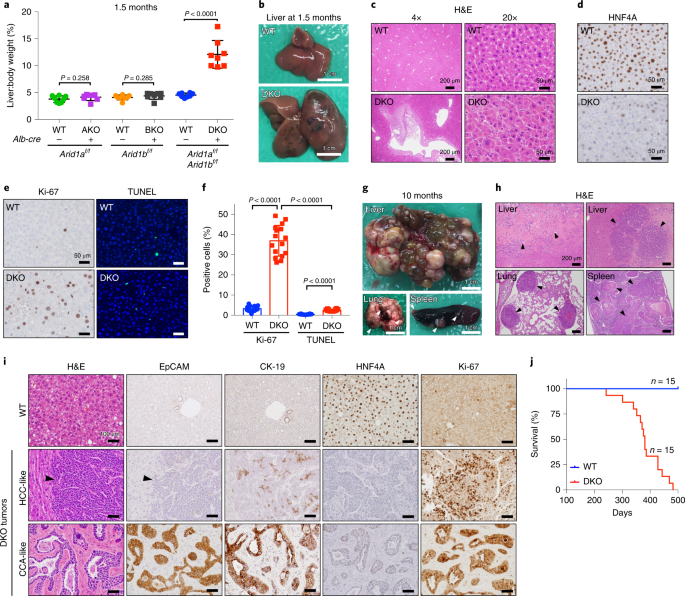
Dual arid1a/arid1b loss leads to rapid carcinogenesis and disruptive redistribution of baf complexes
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT SWI/SNF chromatin remodelers play critical roles in development and cancer. The causal links between SWI/SNF complex disassembly and carcinogenesis are obscured by redundancy
between paralogous components. Canonical BAF (cBAF)-specific paralogs ARID1A and ARID1B are synthetic lethal in some contexts, but simultaneous mutations in both ARID1s are prevalent in
cancer. To understand if and how cBAF abrogation causes cancer, we examined the physiological and biochemical consequences of ARID1A/ARID1B loss. In double-knockout liver and skin,
aggressive carcinogenesis followed dedifferentiation and hyperproliferation. In double-mutant endometrial cancer, add-back of either induced senescence. Biochemically, residual cBAF
subcomplexes resulting from loss of ARID1 scaffolding were unexpectedly found to disrupt a polybromo-containing BAF (pBAF) function. Of 69 mutations in the conserved scaffolding domains of
ARID1 proteins observed in human cancer, 37 caused complex disassembly, partially explaining their mutation spectra. ARID1-less, cBAF-less states promote carcinogenesis across tissues, and
suggest caution against paralog-directed therapies for ARID1-mutant cancer. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel
any time Learn more Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS _ARID1A_ DEFICIENCY WEAKENS BRG1-RAD21 INTERACTION THAT JEOPARDIZES CHROMATIN
COMPACTNESS AND DRIVES LIVER CANCER CELL METASTASIS Article Open access 23 October 2021 GENOMIC PROFILING OF THE TRANSCRIPTION FACTOR ZFP148 AND ITS IMPACT ON THE P53 PATHWAY Article Open
access 25 August 2020 _ARID2_ DEFICIENCY PROMOTES TUMOR PROGRESSION AND IS ASSOCIATED WITH HIGHER SENSITIVITY TO CHEMOTHERAPY IN LUNG CANCER Article 19 March 2021 DATA AVAILABILITY All
sequencing data have been deposited in the Gene Expression Omnibus with the accession nos. GSE147664 for mRNA-seq and GSE140183 for ChIP–seq. Source data are provided with this paper. All
other data supporting the findings of the present study are available from the corresponding author on reasonable request. REFERENCES * Kadoch, C. & Crabtree, G. R. Mammalian SWI/SNF
chromatin remodeling complexes and cancer: mechanistic insights gained from human genomics. _Sci. Adv._ 1, e1500447 (2015). Article PubMed PubMed Central CAS Google Scholar * Hodges,
C., Kirkland, J. G. & Crabtree, G. R. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. _Cold Spring Harb. Perspect. Med._ 6, a026930 (2016). Article PubMed PubMed Central
CAS Google Scholar * Ronan, J. L., Wu, W. & Crabtree, G. R. From neural development to cognition: unexpected roles for chromatin. _Nat. Rev. Genet._ 14, 347–359 (2013). Article CAS
PubMed PubMed Central Google Scholar * Son, E. Y. & Crabtree, G. R. The role of BAF (mSWI/SNF) complexes in mammalian neural development. _Am. J. Med. Genet. C Semin. Med. Genet_.
166C, 333–349 (2014). Article PubMed CAS Google Scholar * Staahl, B. T. & Crabtree, G. R. Creating a neural specific chromatin landscape by npBAF and nBAF complexes. _Curr. Opin.
Neurobiol._ 23, 903–913 (2013). Article CAS PubMed Google Scholar * Wang, W. et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. _EMBO J._ 15, 5370–5382
(1996). Article CAS PubMed PubMed Central Google Scholar * Gatchalian, J. et al. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse
embryonic stem cells. _Nat. Commun._ 9, 5139 (2018). Article PubMed PubMed Central CAS Google Scholar * Wang, X. et al. BRD9 defines a SWI/SNF sub-complex and constitutes a specific
vulnerability in malignant rhabdoid tumors. _Nat. Commun._ 10, 1881 (2019). Article PubMed PubMed Central CAS Google Scholar * Mashtalir, N. et al. Modular organization and assembly of
SWI/SNF family chromatin remodeling complexes. _Cell_ 175, 1272–1288 e1220 (2018). Article CAS PubMed PubMed Central Google Scholar * Raab, J. R., Resnick, S. & Magnuson, T.
Genome-wide transcriptional regulation mediated by biochemically distinct swi/snf complexes. _PLoS Genet._ 11, e1005748 (2015). Article PubMed PubMed Central CAS Google Scholar *
Kadoch, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. _Nat. Genet._ 45, 592–601 (2013). Article CAS PubMed
PubMed Central Google Scholar * Biegel, J. A., Busse, T. M. & Weissman, B. E. SWI/SNF chromatin remodeling complexes and cancer. _Am. J. Med. Genet. C Semin. Med. Genet._ 166C, 350–366
(2014). Article PubMed CAS Google Scholar * Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. _Science_ 330, 228–231 (2010).
Article CAS PubMed PubMed Central Google Scholar * Mathur, R. et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. _Nat. Genet._ 49, 296–302
(2017). Article CAS PubMed Google Scholar * Sun, X. et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. _Cancer Cell_ 33, 151–152 (2018).
Article CAS PubMed PubMed Central Google Scholar * Bitler, B. G. et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. _Nat. Med._ 21,
231–238 (2015). Article CAS PubMed Google Scholar * Fukumoto, T. et al. Repurposing pan-HDAC Inhibitors for ARID1A-mutated ovarian cancer. _Cell Rep._ 22, 3393–3400 (2018). Article CAS
PubMed PubMed Central Google Scholar * Lissanu Deribe, Y. et al. Mutations in the SWI/SNF complex induce a targetable dependence on oxidative phosphorylation in lung cancer. _Nat. Med._
24, 1047–1057 (2018). Article CAS PubMed Google Scholar * McDonald, E. R. 3rd et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by
large-scale, deep RNAi screening. _Cell_ 170, 577–592 e510 (2017). Article CAS PubMed Google Scholar * Michel, B. C. et al. A non-canonical SWI/SNF complex is a synthetic lethal target
in cancers driven by BAF complex perturbation. _Nat. Cell Biol._ 20, 1410–1420 (2018). Article CAS PubMed PubMed Central Google Scholar * Ogiwara, H. et al. Targeting the vulnerability
of glutathione metabolism in ARID1A-deficient cancers. _Cancer Cell_ 35, 177–190 e178 (2019). Article CAS PubMed Google Scholar * Williamson, C. T. et al. ATR inhibitors as a synthetic
lethal therapy for tumours deficient in ARID1A. _Nat. Commun._ 7, 13837 (2016). Article CAS PubMed PubMed Central Google Scholar * Wu, C. et al. Targeting AURKA-CDC25C axis to induce
synthetic lethality in ARID1A-deficient colorectal cancer cells. _Nat. Commun._ 9, 3212 (2018). Article PubMed PubMed Central CAS Google Scholar * Shen, J. et al. ARID1A deficiency
promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. _Nat. Med._ 24, 556–562 (2018). Article CAS PubMed PubMed Central Google
Scholar * Hoffman, G. R. et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. _Proc. Natl Acad. Sci. USA_ 111,
3128–3133 (2014). Article CAS PubMed PubMed Central Google Scholar * Viswanathan, S. R. et al. Genome-scale analysis identifies paralog lethality as a vulnerability of chromosome 1p
loss in cancer. _Nat. Genet._ 50, 937–943 (2018). Article CAS PubMed PubMed Central Google Scholar * Helming, K. C. et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers.
_Nat. Med._ 20, 251–254 (2014). Article CAS PubMed PubMed Central Google Scholar * Kelso, T. W. R. et al. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in
ARID1A-mutant cancers. _eLife._ 6, e30506 (2017). Article PubMed PubMed Central Google Scholar * Coatham, M. et al. Concurrent ARID1A and ARID1B inactivation in endometrial and ovarian
dedifferentiated carcinomas. _Mod. Pathol._ 29, 1586–1593 (2016). Article CAS PubMed Google Scholar * Sun, X. et al. Suppression of the SWI/SNF component Arid1a Promotes mammalian
regeneration. _Cell Stem Cell_ 18, 456–466 (2016). Article CAS PubMed PubMed Central Google Scholar * Celen, C. et al. Arid1b haploinsufficient mice reveal neuropsychiatric phenotypes
and reversible causes of growth impairment. _eLife_ 6, e25730 (2017). Article PubMed PubMed Central Google Scholar * Oba, A. et al. ARID2 modulates DNA damage response in human
hepatocellular carcinoma cells. _J. Hepatol._ 66, 942–951 (2017). Article CAS PubMed Google Scholar * Zhao, H. et al. ARID2: a new tumor suppressor gene in hepatocellular carcinoma.
_Oncotarget_ 2, 886–891 (2011). Article PubMed PubMed Central Google Scholar * Jiang, H. et al. Chromatin remodeling factor ARID2 suppresses hepatocellular carcinoma metastasis via
DNMT1-Snail axis. _Proc. Natl Acad. Sci. USA_ 117, 4770–4780 (2020). Article CAS PubMed PubMed Central Google Scholar * Li, M. et al. Inactivating mutations of the chromatin remodeling
gene ARID2 in hepatocellular carcinoma. _Nat. Genet._ 43, 828–829 (2011). Article CAS PubMed PubMed Central Google Scholar * Kirmitzoglou, I. & Promponas, V. J. LCR-eXXXplorer: a
web platform to search, visualize and share data for low complexity regions in protein sequences. _Bioinformatics_ 31, 2208–2210 (2015). Article CAS PubMed PubMed Central Google Scholar
* Santen, G. W. et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin–Siris syndrome. _Nat. Genet._ 44, 379–380 (2012). Article CAS PubMed Google Scholar *
Wang, K. et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. _Nat. Genet._ 43, 1219–1223 (2011). Article CAS PubMed Google Scholar *
Tsurusaki, Y. et al. Mutations affecting components of the SWI/SNF complex cause Coffin–Siris syndrome. _Nat. Genet._ 44, 376–378 (2012). Article CAS PubMed Google Scholar * Sausen, M.
et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. _Nat. Genet._ 45, 12–17 (2013). Article CAS PubMed Google Scholar * Jiao,
Y. et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. _Nat. Genet._ 45, 1470–1473 (2013). Article CAS PubMed
PubMed Central Google Scholar * Holz-Schietinger, C., Matje, D. M., Harrison, M. F. & Reich, N. O. Oligomerization of DNMT3A controls the mechanism of de novo DNA methylation. _J.
Biol. Chem._ 286, 41479–41488 (2011). Article CAS PubMed PubMed Central Google Scholar * Jurkowska, R. Z. et al. Oligomerization and binding of the Dnmt3a DNA methyltransferase to
parallel DNA molecules: heterochromatic localization and role of Dnmt3L. _J. Biol. Chem._ 286, 24200–24207 (2011). Article CAS PubMed PubMed Central Google Scholar * Nemeth, A.,
Guibert, S., Tiwari, V. K., Ohlsson, R. & Langst, G. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. _EMBO J._ 27, 1255–1265 (2008). Article CAS
PubMed PubMed Central Google Scholar * Zhu, H. et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. _Nat. Genet._ 42,
626–630 (2010). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank C. Kadoch, S. McBrayer, J. Wu, J. Xu, L. Banaszynski, X. Liu and S. Wang
for constructive comments on the manuscript; C. Lewis and J. Shelton for histopathology; Proteomics Core at UTSW (A. Lemoff) for MS; and the CRI Sequencing Core (J. Xu) for genomics. Funding
sources: NIH R03ES026397-01 (to T.W.), CPRIT RP150596 (to T.W.), CPRIT RP170267 (to H.Z.), NIH/NIDDK R01DK111588 (to H.Z.) and Stand Up To Cancer Innovative Research Grant (no.
SU2C-AACR-IRG 10-16 to H.Z.). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Children’s Research Institute, Departments of Pediatrics and Internal Medicine, Center for Regenerative Science
and Medicine, University of Texas Southwestern Medical Center, Dallas, TX, USA Zixi Wang, Yuemeng Jia, Jen-Chieh Chuang, Xuxu Sun, Yu-Hsuan Lin, Cemre Celen, Lin Li, Fang Huang, Xin Liu
& Hao Zhu * Quantitative Biomedical Research Center, Department of Population and Data Sciences, University of Texas Southwestern Medical Center, Dallas, TX, USA Kenian Chen & Tao
Wang * Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX, USA Diego H. Castrillon Authors * Zixi Wang View author publications You can also search for this
author inPubMed Google Scholar * Kenian Chen View author publications You can also search for this author inPubMed Google Scholar * Yuemeng Jia View author publications You can also search
for this author inPubMed Google Scholar * Jen-Chieh Chuang View author publications You can also search for this author inPubMed Google Scholar * Xuxu Sun View author publications You can
also search for this author inPubMed Google Scholar * Yu-Hsuan Lin View author publications You can also search for this author inPubMed Google Scholar * Cemre Celen View author publications
You can also search for this author inPubMed Google Scholar * Lin Li View author publications You can also search for this author inPubMed Google Scholar * Fang Huang View author
publications You can also search for this author inPubMed Google Scholar * Xin Liu View author publications You can also search for this author inPubMed Google Scholar * Diego H. Castrillon
View author publications You can also search for this author inPubMed Google Scholar * Tao Wang View author publications You can also search for this author inPubMed Google Scholar * Hao Zhu
View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Z.W. and H.Z. conceived the project, performed the experiments and wrote the manuscript.
J-C.C., X.S., Z.W., L.L. and C.C. created and analyzed the mouse models. K.C., Y.J., X.S., F.H., X.L. and T.W. generated and analyzed genomic data. Y.-H.L. assisted with the histology
analysis. D.H.C edited the manuscript and provided assistance with disease models. CORRESPONDING AUTHOR Correspondence to Hao Zhu. ETHICS DECLARATIONS COMPETING INTERESTS At the time of
publication, H.Z. owned Ionis Pharmaceuticals stock worth less than $US10,000 and has active collaboration with Alnylam Pharmaceuticals and Twenty-Eight Seven Therapeutics. The remaining
authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 LOSS OF BOTH ARID1A AND ARID1B IN THE LIVER LEADS TO IMPAIRED LIVER FUNCTION. A Kaplan-Meier survival curve of WT and DKO mice within the
first month of life. B Body weight of WT and DKO mice at the age of 1 month (n = 12 and 8 mice). c-g. Liver function analysis using plasma (n = 6 and 5 mice for the _Arid1a__f/f_ group; 6
and 6 for the _Arid1b__f/f_ group; 8 and 8 for the _Arid1a__f/f__; Arid1b__f/f_ group). H Gross inspection of plasma from AKO, BKO and DKO and their corresponding WT mice. I IHC staining of
ARID1A and ARID1B on WT and DKO liver sections. J Western blot showing ARID1A and ARID1B protein levels in WT and DKO livers. K Quantification of Western blot data in J (n = 6 and 6 mice for
each group). Data are presented as mean ± s.d. (B-G,K). Statistical significance was determined by two-tailed unpaired Student’s t-tests with Welch’s correction (B-G, K). Source data
EXTENDED DATA FIG. 2 AAV MEDIATED DELETION OF ARID1A AND ARID1B IN THE LIVER LEADS TO ORGAN FAILURE. A Gross inspection of plasma from mice injected with AAV-GFP or AAV-Cre. B-F. Liver
function tests of _Arid1a__f/f__; Arid1b__f/f_ mice injected with AAV-GFP or AAV-Cre (n = 10 and 10 mice for AST; 9 and 10 for ALT; 9 and 11 for TBIL; 10 and 11 for ALKP; 9 and 11 for
Albumin. Data are presented as mean ± s.d. Statistical significance was determined by two-tailed unpaired Student’s t-tests with Welch’s correction). G Representative genome browser tracks
showing ARID1A binding to the promoter or enhancer regions of differentiation and Cytochrome P450 genes in liver. H IHC staining of EpCAM and CK-19 on AAV-Cre liver sections. Source data
EXTENDED DATA FIG. 3 CBAF SUBUNIT LEVELS SHOWED LIMITED TO NO DECREASE IN ARID1-LESS CELLS OR DKO LIVERS. A Western blot analysis of cBAF subunit levels in WT and ARID1-less H2.35 cells (n =
3 and 3 independent clones). B Colony formation assay for control and ARID1-less H2.35 cells. 0.1 million H2.35 cells were seeded in each well of 6-well plate and cultured for 10 days in
the presence of Dox. C Western blot analysis of cBAF subunit levels in WT and DKO livers (n = 6 and 6 mice). Same batch of western blots/protein samples as in Extended Data Fig. 1j. D
Quantification of western blot data in c (Data are presented as mean ± s.d. Statistical significance was determined by two-tailed unpaired Student’s t-tests with Welch’s correction). Source
data EXTENDED DATA FIG. 4 CHIP-SEQ ANALYSIS OF SWI/SNF COMPLEXES BINDING TO GENOMIC DNA IN CONTROL AND ARID1-LESS H2.35 CELLS. A Expression of Ty1 tagged BRD9 and Brg1 in WT and ARID1-less
H2.35 cells. BRD9 expression was only examined using the Ty1 antibody due to the lack of a commercial anti-mouse BRD9 antibody. B Heatmap displaying ChIP-seq peaks of intact SWI/SNF
complexes in WT H2.35 cells. ARID1A, ARID1B, and BAF45d peaks were used to represent cBAF, ARID2 for pBAF, BRD9 for ncBAF, and Brg1 for all BAF complexes. 3000 bp upstream and downstream of
peak centers are shown in this figure (n = 2 independent ChIP experiments for each protein). C Venn diagram showing the shared and unique binding loci among three types of BAF complexes from
ChIP-seq data. D Comparison of BRD9 occupancies in control and ARID1-less cells. Heatmap and the corresponding averaged peak map and Venn diagram are shown (n = 2 and 2 independent ChIP
experiments). E Representative genome browser tracks showing that ncBAF binding was unaffected in ARID1-less cells (BRD9 peaks in ARID1-less cells). Source data EXTENDED DATA FIG. 5 MAPPING
OF DOMAINS, RESIDUES, AND MUTATIONS RESPONSIBLE FOR ARID1A’S SCAFFOLDING ROLE. A Multiple sequence alignment of ARID1A protein C-terminal regions from human, mouse, dog, bovine, rabbit,
chicken, clawed frog, and zebrafish showing two conserved ARID1 scaffolding domains (ASD1 and ASD2). B Secondary structure prediction of ARID1A using LCR-eXXXplorer server. Regions with a
score lower than 0.5 (shown as a cyan line for the IUPRED score and a red line for the ANCHOR score) are likely well-folded globular domains. C Alanine scans within ASD1 of ARID1A and IP
experiments to assess residues for cBAF subunit interactions. The indicated two residues were mutated to alanine in each construct. D IP experiments showing the influence of ARID1A missense
mutations within ASD2 on BAF subunit interactions. E Western blot showing the influence of ARID1A hotspot missense mutations on protein stability in H2.35 cells. F Western blot showing the
influence of ARID1A truncations on protein stability in H2.35 cells. Source data SUPPLEMENTARY INFORMATION REPORTING SUMMARY SUPPLEMENTARY TABLE Supplementary Tables 1 and 2. SOURCE DATA
SOURCE DATA FIG. 1 Statistical source data. SOURCE DATA FIG. 2 Unprocessed western blots. SOURCE DATA FIG. 3 Statistical source data. SOURCE DATA FIG. 5 Statistical source data. SOURCE DATA
FIG. 5 Unprocessed western blots. SOURCE DATA FIG. 6 Statistical source data. SOURCE DATA FIG. 6 Unprocessed western blots. SOURCE DATA FIG. 7 Statistical source data. SOURCE DATA FIG. 7
Unprocessed western blots. SOURCE DATA FIG. 8 Unprocessed western blots. SOURCE DATA EXTENDED DATA FIG. 1 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 1 Unprocessed western blots.
SOURCE DATA EXTENDED DATA FIG. 2 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 3 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 3 Unprocessed western blots. SOURCE DATA
EXTENDED DATA FIG. 4 Unprocessed western blots. SOURCE DATA EXTENDED DATA FIG. 5 Unprocessed western blots. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Wang, Z., Chen, K., Jia, Y. _et al._ Dual ARID1A/ARID1B loss leads to rapid carcinogenesis and disruptive redistribution of BAF complexes. _Nat Cancer_ 1, 909–922 (2020).
https://doi.org/10.1038/s43018-020-00109-0 Download citation * Received: 17 October 2019 * Accepted: 03 August 2020 * Published: 07 September 2020 * Issue Date: September 2020 * DOI:
https://doi.org/10.1038/s43018-020-00109-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
