Loss-of-function mutations in dnmt3a and tet2 lead to accelerated atherosclerosis and concordant macrophage phenotypes
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Clonal hematopoiesis of indeterminate potential (CHIP) is defined by the presence of a cancer-associated somatic mutation in white blood cells in the absence of overt hematological
malignancy. It arises most commonly from loss-of-function mutations in the epigenetic regulators _DNMT3A_ and _TET2_. CHIP predisposes to both hematological malignancies and atherosclerotic
cardiovascular disease in humans. Here we demonstrate that loss of _Dnmt3a_ in myeloid cells increased murine atherosclerosis to a similar degree as previously seen with loss of _Tet2_. Loss
of _Dnmt3a_ enhanced inflammation in macrophages in vitro and generated a distinct adventitial macrophage population in vivo which merges a resident macrophage profile with an inflammatory
cytokine signature. These changes surprisingly phenocopy the effect of loss of _Tet2_. Our results identify a common pathway promoting heightened innate immune cell activation with loss of
either gene, providing a biological basis for the excess atherosclerotic disease burden in carriers of these two most prevalent CHIP mutations. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 digital issues and online
access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local
taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING
VIEWED BY OTHERS CLONAL HEMATOPOIESIS-RELATED MUTANT ASXL1 PROMOTES ATHEROSCLEROSIS IN MICE VIA DYSREGULATED INNATE IMMUNITY Article 09 December 2024 _TP53-_MEDIATED CLONAL HEMATOPOIESIS
CONFERS INCREASED RISK FOR INCIDENT ATHEROSCLEROTIC DISEASE Article 16 January 2023 THE AIM2 INFLAMMASOME EXACERBATES ATHEROSCLEROSIS IN CLONAL HAEMATOPOIESIS Article 17 March 2021 DATA
AVAILABILITY The data supporting the findings of the present study are available within the paper and Supplementary information and Source data files. All sequencing data from the present
study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus, accession nos. GSE225773 for scRNA-seq and GSE237599 for bulk RNA-seq. The UCSC mm10
reference genome was used for alignment of sequencing data. Multiplexed imaging data has been deposited in Mendeley (https://data.mendeley.com/datasets/dgyrt473vs/1). CODE AVAILABILITY R
code used to analyze scRNA-seq data has been deposited in GitHub (https://github.com/jkgopa/DNMT3A-single-cell). REFERENCES * Jaiswal, S. & Ebert, B. L. Clonal hematopoiesis in human
aging and disease. _Science_ 366, eaan4673 (2019). Article CAS PubMed PubMed Central Google Scholar * Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse
outcomes. _N. Engl. J. Med._ 371, 2488–2498 (2014). Article PubMed PubMed Central Google Scholar * Nowbar, A. N., Gitto, M., Howard, J. P., Francis, D. P. & Al-Lamee, R. Mortality
from ischemic heart disease. _Circ Cardiovasc. Qual. Outcomes_ 12, e005375 (2019). Article PubMed PubMed Central Google Scholar * Smith, Z. D. & Meissner, A. DNA methylation: roles
in mammalian development. _Nat. Rev. Genet._ 14, 204–220 (2013). Article CAS PubMed Google Scholar * Bowman, R. L., Busque, L. & Levine, R. L. Clonal hematopoiesis and evolution to
hematopoietic malignancies. _Cell Stem Cell_ 22, 157–170 (2018). Article CAS PubMed PubMed Central Google Scholar * Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion,
ES-cell self-renewal and inner cell mass specification. _Nature_ 466, 1129–1133 (2010). Article CAS PubMed PubMed Central Google Scholar * Ito, S. et al. Tet proteins can convert
5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. _Science_ 333, 1300–1303 (2011). Article CAS PubMed PubMed Central Google Scholar * Moran-Crusio, K. et al. Tet2 loss leads
to increased hematopoietic stem cell self-renewal and myeloid transformation. _Cancer Cell_ 20, 11–24 (2011). Article CAS PubMed PubMed Central Google Scholar * Challen, G. A. et al.
Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. _Cell Stem Cell_ 15, 350–364 (2014). Article CAS PubMed PubMed Central Google Scholar * Cimmino,
L. et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. _Cell_ 170, 1079–1095.e20 (2017). * Pan, W. et al. The DNA methylcytosine dioxygenase Tet2
sustains immunosuppressive function of tumor-infiltrating myeloid cells to promote melanoma progression. _Immunity_ 47, 284–297.e5 (2017). * Li, Z. et al. Deletion of Tet2 in mice leads to
dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. _Blood_ 118, 4509–4518 (2011). Article CAS PubMed PubMed Central Google Scholar * Jaiswal, S.
et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. _N. Engl. J. Med._ 377, 111–121 (2017). Article PubMed PubMed Central Google Scholar * Bick, A. G. et al.
Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. _Circulation_ 141, 124–131 (2020). Article CAS PubMed Google Scholar * Sano, S. et al.
CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. _Circ. Res._ 123, 335–341 (2018). Article CAS PubMed PubMed Central
Google Scholar * Dorsheimer, L. et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. _JAMA Cardiol_. 4, 25–33 (2019).
Article PubMed Google Scholar * Fuster, J. J. et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. _Science_ 355, 842–847 (2017).
Article CAS PubMed PubMed Central Google Scholar * Robbins, C. S. et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. _Nat. Med._ 19, 1166–1172
(2013). Article CAS PubMed PubMed Central Google Scholar * Williams, J. W. et al. Limited proliferation capacity of aortic intima resident macrophages requires monocyte recruitment for
atherosclerotic plaque progression. _Nat. Immunol._ 21, 1194–1204 (2020). Article CAS PubMed PubMed Central Google Scholar * Swirski, F. K. et al. Ly-6Chi monocytes dominate
hypercholesterolemia-associated monocytosis and give rise to macrophages in atheromata. _J. Clin. Invest._ 117, 195–205 (2007). Article CAS PubMed PubMed Central Google Scholar *
Svensson, E. C. et al. TET2-driven clonal hematopoiesis and response to canakinumab: an exploratory analysis of the CANTOS randomized clinical trial. _JAMA Cardiol._ 7, 521–528 (2022).
Article PubMed PubMed Central Google Scholar * Fidler, T. P. et al. The AIM2 inflammasome exacerbates atherosclerosis in clonal haematopoiesis. _Nature_ 592, 296–301 (2021). Article CAS
PubMed PubMed Central Google Scholar * Libby, P., Lichtman, A. H. & Hansson, G. K. Immune effector mechanisms implicated in atherosclerosis: from mice to humans. _Immunity_ 38,
1092–1104 (2013). Article CAS PubMed PubMed Central Google Scholar * Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data
across different conditions, technologies, and species. _Nat. Biotechnol._ 36, 411–420 (2018). Article CAS PubMed PubMed Central Google Scholar * Stuart, T. et al. Comprehensive
integration of single-cell data. _Cell_ 177, 1888–1902.e21 (2019). * Zernecke, A. et al. Meta-analysis of leukocyte diversity in atherosclerotic mouse aortas. _Circ. Res._ 127, 402–426
(2020). Article CAS PubMed PubMed Central Google Scholar * Cochain, C. et al. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine
atherosclerosis. _Circ. Res._ 122, 1661–1674 (2018). * Winkels, H. et al. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass
cytometry. _Circ. Res._ 122, 1675–1688 (2018). Article CAS PubMed PubMed Central Google Scholar * Tacke, F. et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to
accumulate within atherosclerotic plaques. _J. Clin. Invest._ 117, 185–194 (2007). Article CAS PubMed PubMed Central Google Scholar * Keren, L. et al. A structured tumor-immune
microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. _Cell_ 174, 1373–1387.e19 (2018). * Epelman, S. et al. Embryonic and adult-derived resident
cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. _Immunity_ 40, 91–104 (2014). Article CAS PubMed PubMed Central Google Scholar *
Libby, P. The changing landscape of atherosclerosis. _Nature_ 592, 524–533 (2021). Article CAS PubMed Google Scholar * Kim, P. G. et al. Dnmt3a-mutated clonal hematopoiesis promotes
osteoporosis. _J. Exp. Med._ 218, e20211872 (2021). Article CAS PubMed PubMed Central Google Scholar * Nahrendorf, M. Myeloid cell contributions to cardiovascular health and disease.
_Nat. Med._ 24, 711–720 (2018). Article CAS PubMed PubMed Central Google Scholar * Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. _N. Engl.
J. Med._ 377, 1119–1131 (2017). Article CAS PubMed Google Scholar * Wu, H. et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in
transcriptional regulation in mouse embryonic stem cells. _Genes Dev._ 25, 679–684 (2011). Article CAS PubMed PubMed Central Google Scholar * Cobo, I. et al. DNA methyltransferase 3
alpha and TET methylcytosine dioxygenase 2 restrain mitochondrial DNA-mediated interferon signaling in macrophages. _Immunity_ 55, 1386–1401.e10 (2022). * Russler-Germain, D. A. et al. The
R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. _Cancer Cell_ 25, 442–454 (2014). Article CAS PubMed
PubMed Central Google Scholar * Kim, S. J. et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. _Blood_ 122, 4086–4089 (2013). Article CAS PubMed
PubMed Central Google Scholar * Huang, Y. H. et al. Systematic profiling of DNMT3A variants reveals protein instability mediated by the DCAF8 E3 ubiquitin ligase adaptor. _Cancer
Discov._ 12, 220–235 (2022). * Yang, L., Rau, R. & Goodell, M. A. DNMT3A in haematological malignancies. _Nat. Rev. Cancer_ 15, 152–165 (2015). Article CAS PubMed PubMed Central
Google Scholar * Saiki, R. et al. Combined landscape of single-nucleotide variants and copy number alterations in clonal hematopoiesis. _Nat. Med._ 27, 1239–1249 (2021). Article CAS
PubMed Google Scholar * Nachun, D. et al. Clonal hematopoiesis associated with epigenetic aging and clinical outcomes. _Aging Cell_ 20, e13366 (2021). Article CAS PubMed PubMed Central
Google Scholar * Mokry, M. et al. Transcriptomic-based clustering of human atherosclerotic plaques identifies subgroups with different underlying biology and clinical presentation. _Nat.
Cardiovasc. Res._ 1, 1140–1155 (2022). Article Google Scholar * Andrysik, Z., Sullivan, K. D., Kieft, J. S. & Espinosa, J. M. PPM1D suppresses p53-dependent transactivation and cell
death by inhibiting the integrated stress response. _Nat. Commun._ 13, 7400 (2022). Article CAS PubMed PubMed Central Google Scholar * Nguyen, S., Meletis, K., Fu, D., Jhaveri, S. &
Jaenisch, R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. _Dev. Dyn._ 236, 1663–1676 (2007). Article CAS
PubMed Google Scholar * Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. _Nucleic Acids Res._ 44, W90–W97 (2016). Article CAS PubMed
PubMed Central Google Scholar * Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. _OMICS_ 16, 284–287 (2012).
Article CAS PubMed PubMed Central Google Scholar * Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at _bioRxiv_ https://doi.org/10.1101/060012 (2021). * Haber, A. L.
et al. A single-cell survey of the small intestinal epithelium. _Nature_ 551, 333–339 (2017). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We
thank O. Rozenblatt-Rosen (of Genentech, previously Broad Institute) and A. Rotem (of AstraZeneca, previously the Dana-Farber Cancer Institute) for expert advice regarding scRNA-seq and A.
Sperling and M. Slabicki (both of the Dana-Farber Cancer Institute) for helpful discussions. We further thank the staff at the Klarman Cell Observatory of the Broad Institute, the Rodent
Histopathology Core at Harvard Medical School, the Stanford Research Computing Center and M. Leventhal for technical assistance. Illustrations were created with BioRender. P.J.R.
acknowledges funding support from the EvansMDS Foundation and the John R. Svenson Endowed Fellowship. T.N. is supported by the Japan Society for the Promotion of Science Overseas Fellowship.
A.E.L. reports funding from the John S. LaDue Memorial Fellowship in Cardiology, the American Society of Hematology Research Training Grant for Fellows and the American College of
Cardiology Merck Fellowship in Cardiovascular Disease and the Metabolic Syndrome. M.A. is supported by funding from the NIH (grant nos. 5U54CA20997105, 5DP5OD01982205, 1R01CA24063801A1,
5R01AG06827902, 5UH3CA24663303, 5R01CA22952904, 1U24CA22430901, 5R01AG05791504 and 5R01AG05628705), the Department of Defense (grant no. W81XWH2110143) and other funding from the Bill and
Malinda Gates Foundation, the Cancer Research Institute, the Parker Center for Cancer Immunotherapy and the Breast Cancer Research Foundation. P.L. receives funding support from the National
Heart, Lung, and Blood Institute (grant nos. 1R01HL134892 and 1R01HL163099-01), the RRM Charitable Fund and the Simard Fund. B.L.E. is an investigator of the Howard Hughes Medical Institute
and received funding from the NIH (grant nos. P01CA066996, P50CA206963 and R01HL082945). S.J. is supported by the Burroughs Wellcome Fund, EvansMDS Foundation, Ludwig Center for Cancer Stem
Cell Research, Leukemia and Lymphoma Society, Knight Initiative for Brain Resilience, the NIH (grant no. DP2-HL157540) and the Leducq Foundation. AUTHOR INFORMATION Author notes * These
authors contributed equally: Philipp J. Rauch, Jayakrishnan Gopakumar. AUTHORS AND AFFILIATIONS * Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA Philipp J.
Rauch, Marie McConkey, Tetsushi Nakao, Amy E. Lin, Drew N. Cohen & Benjamin L. Ebert * Broad Institute of Harvard and MIT, Cambridge, MA, USA Philipp J. Rauch, Marie McConkey, Tetsushi
Nakao, Amy E. Lin & Benjamin L. Ebert * Department of Pathology, Stanford University School of Medicine, Stanford, CA, USA Jayakrishnan Gopakumar, Daniel Nachun, Herra Ahmad, Marc Bosse,
Nora Vivanco Gonzalez, Noah F. Greenwald, Erin F. McCaffrey, Zumana Khair, Kameron B. Rodrigues, Eti Sinha, Sean Bendall, Michael Angelo & Siddhartha Jaiswal * Division of Hematology,
Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA Alexander J. Silver * Cardiovascular Research Center and Center for Genomic Medicine, Massachusetts General Hospital,
Boston, MA, USA Tetsushi Nakao * Cardiovascular Division, Department of Medicine, Brigham and Women’s Hospital, Boston, MA, USA Tetsushi Nakao, Thiago Rentz, Amy E. Lin, Maia Fefer, Amélie
Vromman, Eugenia Shvartz, Galina Sukhova & Peter Libby * Department of Electrical Engineering, Stanford University, Stanford, CA, USA Manu Gopakumar * Howard Hughes Medical Institute,
Boston, MA, USA Benjamin L. Ebert * Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Stanford, CA, USA Siddhartha Jaiswal Authors * Philipp
J. Rauch View author publications You can also search for this author inPubMed Google Scholar * Jayakrishnan Gopakumar View author publications You can also search for this author inPubMed
Google Scholar * Alexander J. Silver View author publications You can also search for this author inPubMed Google Scholar * Daniel Nachun View author publications You can also search for
this author inPubMed Google Scholar * Herra Ahmad View author publications You can also search for this author inPubMed Google Scholar * Marie McConkey View author publications You can also
search for this author inPubMed Google Scholar * Tetsushi Nakao View author publications You can also search for this author inPubMed Google Scholar * Marc Bosse View author publications You
can also search for this author inPubMed Google Scholar * Thiago Rentz View author publications You can also search for this author inPubMed Google Scholar * Nora Vivanco Gonzalez View
author publications You can also search for this author inPubMed Google Scholar * Noah F. Greenwald View author publications You can also search for this author inPubMed Google Scholar *
Erin F. McCaffrey View author publications You can also search for this author inPubMed Google Scholar * Zumana Khair View author publications You can also search for this author inPubMed
Google Scholar * Manu Gopakumar View author publications You can also search for this author inPubMed Google Scholar * Kameron B. Rodrigues View author publications You can also search for
this author inPubMed Google Scholar * Amy E. Lin View author publications You can also search for this author inPubMed Google Scholar * Eti Sinha View author publications You can also search
for this author inPubMed Google Scholar * Maia Fefer View author publications You can also search for this author inPubMed Google Scholar * Drew N. Cohen View author publications You can
also search for this author inPubMed Google Scholar * Amélie Vromman View author publications You can also search for this author inPubMed Google Scholar * Eugenia Shvartz View author
publications You can also search for this author inPubMed Google Scholar * Galina Sukhova View author publications You can also search for this author inPubMed Google Scholar * Sean Bendall
View author publications You can also search for this author inPubMed Google Scholar * Michael Angelo View author publications You can also search for this author inPubMed Google Scholar *
Peter Libby View author publications You can also search for this author inPubMed Google Scholar * Benjamin L. Ebert View author publications You can also search for this author inPubMed
Google Scholar * Siddhartha Jaiswal View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS P.J.R., P.L., B.L.E. and S.J. designed the study.
P.J.R., J.G., A.J.S., H.A., M.M., T.N., T.R., A.E.L., M.F., D.N.C., A.V., E.S., G.S. and S.J. performed and/or analyzed in vivo experiments on atherosclerotic mice, aortic root imaging and
flow cytometry. P.J.R., J.G., A.J.S. and S.J. performed and/or analyzed bulk RNA-seq experiments from BMDMs. P.J.R., J.G., D.N., K.B.R., E.S. and S.J. performed and/or analyzed scRNA-seq
experiments. P.J.R., J.G., M.B., N.V.G., N.F.G, E.F.M., Z.K. and M.G. performed and/or analyzed MIBI-TOF experiments. A.J.S. and S.J. performed and/or analyzed ELISA experiments. S.B. and
M.A. supervised MIBI-TOF experiments. P.L., B.L.E. and S.J. supervised the study. P.J.R. wrote the paper with contributions from all authors. S.J. revised the paper. CORRESPONDING AUTHORS
Correspondence to Philipp J. Rauch or Siddhartha Jaiswal. ETHICS DECLARATIONS COMPETING INTERESTS M.B. is presently a consultant for the company IonPath Inc., which manufactured the MIBI-TOF
instrument used in this paper. A.E.L. is a member of TenSixteen Bio, outside of the submitted work. E.M. previously consulted for IonPath Inc. M.A. is a board member and shareholder in
IonPath, which develops and manufactures the commercial MIBI-TOF platform. P.L. is an unpaid consultant to, or involved in, clinical trials for Amgen, AstraZeneca, Baim Institute, Beren
Therapeutics, Esperion Therapeutics, Genentech, Kancera, Kowa Pharmaceuticals, Medimmune, Merck, Moderna, Novo Nordisk, Novartis, Pfizer and Sanofi-Regeneron. P.L. is a member of the
scientific advisory board for Amgen, Caristo Diagnostics, Cartesian Therapeutics, CSL Behring, DalCor Pharmaceuticals, Dewpoint Therapeutics, Eulicid Bioimaging, Kancera, Kowa
Pharmaceuticals, Olatec Therapeutics, Medimmune, Novartis, PlaqueTec, TenSixteen Bio, Soley Therapeutics and XBiotech, Inc. P.L.’s laboratory has received research funding in the last 2
years from Novartis, Novo Nordisk and Genentech. P.L. is on the Board of Directors of XBiotech, Inc. and has a financial interest in XBiotech, a company developing therapeutic human
antibodies; in TenSixteen Bio, a company targeting somatic mosaicism and CHIP to discover and develop new therapeutics to treat age-related diseases; and in Soley Therapeutics, a
biotechnology company that is combining artificial intelligence with molecular and cellular response detection for discovering and developing new drugs, currently focusing on cancer
therapeutics. P.L.’s interests were reviewed and are managed by Brigham and Women’s Hospital and Mass General Brigham in accordance with their conflict-of-interest policies. B.L.E. has
received consulting fees from GRAIL. He is a member of the scientific advisory board and shareholder for Neomorph Therapeutics, Skyhawk Therapeutics and Exo Therapeutics. B.L.E.’s laboratory
has received research funding in the last 2 years from Novartis and Calico. S.J. is a scientific advisor to AstraZeneca, Novartis, Genentech, AVRO Bio and Foresite Labs, reports speaking
fees from GSK, is an equity holder and on the scientific advisory board for Bitterroot Bio, and is an equity holder, co-founder and on the scientific advisory board of TenSixteen Bio. The
other authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Cardiovascular Research_ thanks Franco Izzo, Alan Tall and the other, anonymous, reviewer(s) for
their contribution to the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and
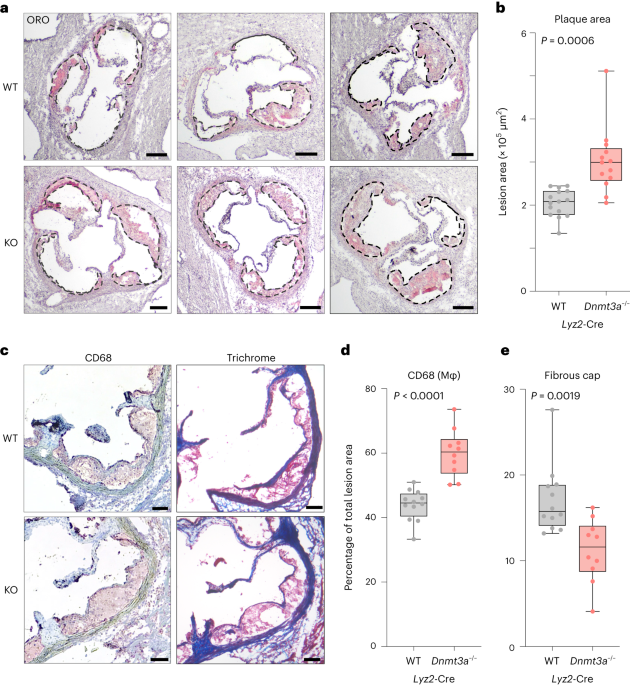
institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 EXTENDED PHENOTYPIC CHARACTERIZATION OF _DNMT3A_ LOSS IMPACTING ATHEROSCLEROSIS. A-B, Bi-allelic loss of _Dnmt3a_ in
hematopoietic cells accelerates atherosclerosis. (A) Oil red O (ORO) stained aortic root sections in female _Ldlr_−_/_− mice transplanted with either _Dnmt3a__+/+_; _Vav1_-Cre (WT) or
_Dnmt3a_−_/_−; _Vav1_-Cre (KO) marrow, in a 1:9 ratio with WT, after 9 weeks of feeding on high-fat, high-cholesterol diet. Atheromata are demarcated by dashed lines. Scale bars = 200 µm.
(B) Quantification of lesion area in the aortic root. N = 10 animals for both groups. Unpaired two-tailed t-test with Welsh’s correction. Box plot shows min, 25th percentile, median, 75th
percentile and max. C, Single-allele loss of _Dnmt3a_ has minimal impact on atherosclerosis. Shown is lesion area after 9 weeks on diet in female _Ldlr_−_/_− mice that were transplanted with
either _Dnmt3a_ + _/−_; _Vav1_-Cre marrow (left) or _Dnmt3a_ + _/−_; _Lyz2_-Cre marrow (right), compared to WT controls. N = 10 animals per group for _Vav1_-Cre experiment, N = 15 per group
for _Lyz2_-cre experiment. Unpaired two-tailed t-test with Welsh’s correction. Box plots show min, 25th percentile, median, 75th percentile and max. D-E, Lesion size in advanced
atherosclerosis does not differ between WT and _Dnmt3a_ knock-out. (D) ORO-stained aortic root sections in female _Ldlr_−_/_− mice transplanted with either _Dnmt3a__+/+_; _Vav1_-Cre (WT) or
_Dnmt3a_−_/_−; _Vav1_-Cre (KO) marrow, after 20 weeks of feeding on high-fat, high-cholesterol diet. Atheromata are demarked by dashed lines. Scale bars = 200 µm. (E) Quantification of
lesional area in the aortic root. N = 4 mice for WT and N = 5 for KO group. Unpaired two-tailed t-test with Welsh’s correction. Box plot shows min, 25th percentile, median, 75th percentile
and max. F, Isotype control (rabbit IgG) IHC staining for CD68 on aortic root with atheroma (control for Fig. 1c-d) demonstrates specificity of staining. Scale bar = 100 µm. EXTENDED DATA
FIG. 2 PERIPHERAL BLOOD COUNTS, CHIMERISM AND SERUM LIPIDS IN MICE TRANSPLANTED WITH _DNMT3A-_DEFICIENT MARROW. A, Peripheral blood chimerism in mice transplanted with either CD45.2+
_Vav1_-Cre (wt) or CD45.2+ _Dnmt3a_−_/_−; _Vav1_-Cre (KO) marrow, in a 1:9 ratio with CD45.1+ WT, after 9 weeks on diet. N = 16 animals for WT group and N = 12 animals for KO group.
Two-tailed unpaired t test was performed to compare the myeloid cell population between groups. B, Peripheral blood cell counts in transplanted mice. Indicated are transplanted genotypes and
time on high-fat, high-cholesterol diet. Groups that received only wild-type marrow are marked in gray, and groups that received marrow with bi-allelic loss of _Dnmt3a_ are marked in red. N
= 9 for wt Vav 8 Wk, N = 11 for KO Vav 8 Wk, N = 12 for wt Vav 20 Wk and KO Vav 20 Wk, N = 13 for wt LysM 8 Wk, N = 15 for KO LysM 8 Wk. P values obtained by two-tailed unpaired t tests. C,
Serum lipid concentrations in mice transplanted with either _Vav1_-Cre (wt) or CD45.2+ _Dnmt3a_−_/_−; _Vav1_-Cre (KO) marrow, after 14 weeks on diet. For total cholesterol, N = 9 for wt, N
= 6 for KO. For HDL, N = 9 for wt, N = 7 for KO. For LDL, N = 9 for wt, N = 6 for KO. For triglycerides, N = 8 for wt, N = 7 for KO. Varying group sizes due to insufficient volume for all
analyses in some samples. All box plots show min, 25th percentile, median, 75th percentile and max. EXTENDED DATA FIG. 3 COMPARATIVE GENE-SET ENRICHMENT ANALYSIS. A, Comparative GSEA using
the merge_result function in clusterProfiler between _Dnmt3a_−_/_− and _Tet2_−_/_−; _Vav1_-Cre BMDM. Top enriched KEGG pathways in _Dnmt3a_−_/_− and _Tet2_−_/_− (showCategory = 10) are
shown. P value estimation in the fgsea method used by clusterProfiler is based on an adaptive multi-level split Monte-Carlo scheme. Adjustment for multiple comparisons was performed using
the Benjamini-Hochberg method. B, Venn diagram showing overlap in the identity of significantly enriched (FDR < 0.05, ES > 0) 2019 KEGG pathways obtained by gene-set enrichment
analysis (GSEA) in _Dnmt3a_−_/_− vs. WT (red) or _Tet2_−/− vs. WT (blue) BMDM, see Supplementary Table for detailed listing of all enriched pathways. Statistical significance calculated by
the hypergeometric test. C, Specificity test of enriched gene distribution analysis using _Jak2_VF as a comparator. Enriched (log2FC > 0.5 and p < 0.05, red dots) or depleted (log2FC
< −0.5 and p < 0.05, green dots) transcripts in _Dnmt3a_−/− BMDM were tested for enrichment or depletion (p < 0.05) in _Jak2_VF BMDM. The resulting distribution for each permutation
was statistically compared to an equipartition (boxes) by way of a two-sided Chi-square test. EXTENDED DATA FIG. 4 CONVERGENT CHANGES IN INFLAMMATORY PATHWAYS WITH _DNMT3A_ AND _TET2_
DEFICIENCY. A, BMDM were cultured with LDL or vehicle (NT = non-treated) for 24 h, and mRNA was assessed by RNA-sequencing. Out of the top 200 genes that were affected by both LDL treatment
and genotype (log2FC > 0.6, adjusted p-value < 0.05), 37 selected genes involved in inflammation are shown. Each column in the heatmap represent an individual biological replicate. B,
Scatter plot comparing gene expression changes in _Dnmt3a_−/− vs. WT with the _Tet2_−/− vs. WT dataset from Jaiswal et al., 2017. Displayed are the most highly expressed genes significantly
affected by 200 mg/dL LDL stimulation in BMDM. Red dots highlight genes involved in chemokine signaling. Shown are mean fold changes over 3 biological replicates per genotype and stimulation
status for the WT vs _Dnmt3a_−/− comparison; for the WT vs. _Tet2_−/− comparison, 3 biological replicates per genotype for the non-treated condition and 2 replicates for the LDL condition.
C, Intracellular flow cytometry validates increase in pro-IL-1β in stimulated _Dnmt3a_−/− and _Tet2_−/− BMDM vs. WT at the protein level. Flow cytometry plot depicts macrophage gating
strategy (showing WT as a representative example). Histograms show unstimulated BMDM (gray) overlaid onto stimulated BMDM (orange). Bar graph shows quantification. N = 3 per group, mean ±
SD. P values obtained by one-way ANOVA followed by Tukey’s post hoc test. D, ELISA for key secreted cytokines measured in supernatant of LDL-stimulated BMDM from WT or _Dnmt3a_−/−. N = 6 per
group (biological replicates). Two-tailed unpaired t-tests. Box plots show min, 25th percentile, median, 75th percentile and max. EXTENDED DATA FIG. 5 SINGLE-CELL RNA SEQUENCING (SCRNA-SEQ)
FROM ATHEROMATA. A, Flow cytometric analysis of chimerism in aortic single cell suspensions from mixed chimeric _Ldlr_−/− mice on 30 weeks of high fat, high cholesterol diet. Transplanted
genotypes and ratios are indicated. Dot plots show live, doublet excluded, CD45 positive cells. B, Violin plots depict single-cell expression of key signature genes across the 5 classic
mononuclear phagocyte populations. Color code follows Fig. 3b. C-E, Lesional cell distribution is shaped by genotype. (C) Major leukocyte lineages. Chi square test. (D) Monocyte subsets. Chi
square test. (E) Proportions-of-clusters analysis within the lesional lymphocyte compartment. Displayed is log2 fold difference and 95% confidence interval. Permutation test with N = 1000
permutations. Full statistics are reported in Supplementary Table 3. F, Single-cell transcriptome-based cell cycle analysis in macrophages stratified by subset and genotype. EXTENDED DATA
FIG. 6 CHIP GIVES RISE TO A DISTINCT MACROPHAGE POPULATION IN ATHEROMATA. A, UMAP plot of highly expressed genes that are differentially upregulated in the _Folr2_+ _Mrc1_+ _Ccl8_+ _Cxcl1_+
macrophage cluster separated by genotype (WT, _Dnmt3a_−_/_−; _Vav1_-Cre, and _Tet2_−_/_−; _Vav1_-Cre). B, Significantly enriched 2019 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
involved in inflammation and aging in the CHIP-TR macrophage population, analogous to the analysis in BMDM depicted in Fig. 2. Tested genes were present in at least 25% of the CHIP-TR
macrophage subpopulation with a positive log fold change of at least 0.25. Dotted blue line indicates P value of 0.05. P values were calculated by Fisher’s exact test using the R package
enrichr. C, GSEA of a pathway comprising the top 25 upregulated genes in _Tet2_−_/_− vs. WT by log2FC within the Resident-like Mφ cluster (‘_Tet2_ KO sc-pathway’) within the _Dnmt3a_−/− vs.
WT ranking in the same cluster, analogous to the BMDM analysis in Fig. 2d. P value estimation in the fgsea method is based on an adaptive multi-level split Monte-Carlo scheme. D, Volcano
plot depicts differential gene expression analysis in _Dnmt3a_−/− vs. _Tet2_−/− within the resident-like macrophage (Mφ) cluster using the FindMarkers function in Seurat (logfc.threshold =
0.25) using a negative binomial generalized linear model. Labeled are genes with a |log2FC|>0.5 (vertical lines) and adjusted p-value < 10e-12 (horizontal line). EXTENDED DATA FIG. 7
EXPANSION OF _FOLR2__+_ _MRC1__+_ _CCL8__+_ _CXCL1__+_ MACROPHAGES IN CHIP. A, UMAP plots of aligned gene expression data in single CD45.2+ cells isolated from aortae from WT; _Vav1_-Cre (n
= 5015), _Dnmt3a_−/−; _Vav1_-Cre (n = 4499), and _Tet2_−/−; _Vav1_-Cre (n = 6078) populations (24-week cohort). B, _Folr2__+_ _Mrc1__+_ _Ccl8__+_ _Cxcl1__+_ (CHIP-TR) macrophages highlighted
on the UMAP plots from A following 24 weeks of high-fat, high-cholesterol diet. Gray dots correspond to all other cells. Quantification in Fig. 4d. C, Gating strategy for identification of
donor-derived tissue resident-like (TR) macrophages in atheromata by flow cytometry. The population was defined as CD45.2+ CD11b+ F4/80+ CD206hi/+. Comparison to other macrophages (CD206lo)
confirmed higher expression of LYVE1 and FOLR2, and lower expression of CD9, as expected from scRNA-seq. EXTENDED DATA FIG. 8 MULTIPLEX ION BEAM IMAGING BY TIME-OF-FLIGHT (MIBI-TOF) OF
AORTIC ROOTS IN WT, _DNMT3A__−/−_ AND _TET2__−/−_. A, MIBI-TOF of aortic roots in WT, _Dnmt3a__−/_− and _Tet2_−_/_−. Shown are 3 exemplary antigens (in white) with relevance to
atherosclerosis (von Willebrand factor [VWF], integrin subunit alpha X/CD11c and mannose receptor 1/CD206) out of 27 total markers recorded. DNA (blue) and SMA (magenta) channels are shown
to provide anatomical reference. Scale bars = 200 μm. B, Biologically informed decision tree used to define cell identities (boxes) based on combinations of antigen markers. Black arrows
signify condition fulfilled/marker positive. Red arrows signify that the preceding (combination of) condition(s) has not been fulfilled. & = AND. ∥ = OR. TR ΜΦ = tissue resident-like
macrophage. EXTENDED DATA FIG. 9 CHIP ALTERS THE SPATIAL CELLULAR COMPOSITION OF ATHEROMATA. A, Quantification strategy for adventitial CD206+ TR macrophages in 5-week roots (N = 9). For
each section (biological replicate), the average of 4 ROIs was reported (demarcated by yellow lines). Scale bars = 200 μm. B, Section of the arterial intima (white rectangles), high-power
images below, depicting nascent atheromata (scale bars = 200 μm). C, IHC for CD206 on aortic roots in WT and _Dnmt3a_−/− mice. Graph shows quantification of 3,3′-Diaminobenzidine (DAB)
density, normalized to (arc) length of the section measured. N = 7 mice per WT and N = 9 mice for KO group. Unpaired two-tailed t-test with Welsh’s correction. Box plots show min, 25th
percentile, median, 75th percentile and max. EXTENDED DATA FIG. 10 SUMMARY. Mutations in the epigenetic regulators _Dnmt3a_ or _Tet2_ in hematopoietic cells converge in the emergence of a
distinct macrophage subset in the arterial adventitia (denoted CHIP Resident-like Mφ) that combines surface markers associated with resident-like macrophages (depicted: mannose
receptor/CD206) with a distinct inflammatory chemokine signature. These cells are surrounded by other myeloid cells and clusters of activated endothelium. Overall, lesional (inflammatory)
macrophage content increases, while other immune cell subsets, in particular T lymphocytes, decrease. Collectively, these processes result in increased atherosclerosis. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION Supplementary Figs. 1–4. REPORTING SUMMARY SUPPLEMENTARY TABLES Supplementary Tables 1–6. SUPPLEMENTARY DATA FILE 1 LDL responsive genes. SUPPLEMENTARY
DATA FILE 2 Cluster marker genes. SUPPLEMENTARY DATA FILE 3 Genes enriched in _Dnmt3a_−/− versus WT within the resident-like macrophage cluster (scRNA-seq). SUPPLEMENTARY DATA FILE 4 Genes
enriched in _Tet2_−/− versus WT within the resident-like macrophage cluster (scRNA-seq). SOURCE DATA SOURCE DATA FIG. 1 Statistical source data. SOURCE DATA FIG. 4 Statistical source data.
SOURCE DATA FIG. 5 Statistical source data. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing
agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement
and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Rauch, P.J., Gopakumar, J., Silver, A.J. _et al._ Loss-of-function mutations in _Dnmt3a_ and _Tet2_ lead to
accelerated atherosclerosis and concordant macrophage phenotypes. _Nat Cardiovasc Res_ 2, 805–818 (2023). https://doi.org/10.1038/s44161-023-00326-7 Download citation * Received: 06 October
2021 * Accepted: 27 July 2023 * Published: 04 September 2023 * Issue Date: September 2023 * DOI: https://doi.org/10.1038/s44161-023-00326-7 SHARE THIS ARTICLE Anyone you share the following
link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature
SharedIt content-sharing initiative
