Characterization of the first double-stranded rna bacteriophage infecting pseudomonas aeruginosa
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

ABSTRACT Bacteriophages (phages) are widely distributed in the biosphere and play a key role in modulating microbial ecology in the soil, ocean, and humans. Although the role of DNA
bacteriophages is well described, the biology of RNA bacteriophages is poorly understood. More than 1900 phage genomes are currently deposited in NCBI, but only 6 dsRNA bacteriophages and 12
ssRNA bacteriophages genome sequences are reported. The 6 dsRNA bacteriophages were isolated from legume samples or lakes with _Pseudomonas syringae_ as the host. Here, we report the first
_Pseudomonas aeruginosa_ phage phiYY with a three-segmented dsRNA genome. phiYY was isolated from hospital sewage in China with the clinical _P. aeruginosa_ strain, PAO38, as a host.
Moreover, the dsRNA phage phiYY has a broad host range, which infects 99 out of 233 clinical _P. aeruginosa_ strains isolated from four provinces in China. This work presented a detailed
characterization of the dsRNA bacteriophage infecting _P. aeruginosa._ SIMILAR CONTENT BEING VIEWED BY OTHERS ISOLATION AND CHARACTERIZATION OF A LYTIC BACTERIOPHAGE AGAINST _PSEUDOMONAS
AERUGINOSA_ Article Open access 29 September 2021 ISOLATION, PHENOTYPIC CHARACTERIZATION AND COMPARATIVE GENOMIC ANALYSIS OF 2019SD1, A POLYVALENT ENTEROBACTERIA PHAGE Article Open access 12
November 2021 A NOVEL VIRULENT _LITUNAVIRUS_ PHAGE POSSESSES THERAPEUTIC VALUE AGAINST MULTIDRUG RESISTANT _PSEUDOMONAS AERUGINOSA_ Article Open access 07 December 2022 INTRODUCTION
Bacteriophages (phages) are estimated to be the most abundant organisms on earth, with key roles in shaping the composition of microbial ecology by lysing bacteria or transferring genes1,2.
Despite the numerous available studies on DNA bacteriophages, the biology of RNA bacteriophages has been poorly investigated. Recently, Siddharth _et al_. identified the partial genome
sequences of 122 RNA bacteriophage phylotypes by metagenomic sequencing2. Their results indicated that RNA phages are globally distributed in numerous microbial communities. More than 1900
phage genomes are currently deposited in NCBI. However, only the genome sequences of 6 double-stranded RNA (dsRNA) bacteriophages (phi6, phi8, phi12, phi13, phi2954, and phiNN) and 12
single-stranded RNA (ssRNA) bacteriophages are available in the GenBank Genomes database as of 1 September, 2016 (Supplementary Tables S1 and S2). Therefore, more unrecognized RNA
bacteriophages have yet to be discovered. The discovery of dsRNA phages dates back to 1973, when Vidaver _et al_. identified a lipid-containing bacteriophage, phi6, with a three-segmented
dsRNA genome3. The characterization of these viruses has improved the understanding of dsRNA virus biology, such as its replication, virus assembly, genome packaging, and phage–host
interactions4,5,6,7,8,9. All dsRNA phages are classified in the Cystoviridae family; phi6 is a representative member of Cystoviridae. The three genome segments of phi6 are designated L (6374
bp), M (4063 bp), and S (2948 bp). Currently, more than 10 dsRNA phages have been isolated; 6 of which (phi6, phi8, phi12, phi13, phi2954, and phiNN) have genome sequences deposited in
GenBank10,11,12,13,14,15. We previously isolated several dsDNA phages infecting _Pseudomonas aeruginosa_16,17. Here, we report a dsRNA phage phiYY isolated from the sewage of Southwest
Hospital, with the clinical _P. aeruginosa_ strain, PAO38, as the host. Compared with the previously identified dsRNA phages from legume samples or lakes, phiYY was isolated from hospital
sewage and was the first dsRNA phage known to infect the opportunistic pathogen _P. aeruginosa_. RESULTS AND DISCUSSION BIOLOGICAL CHARACTERISTICS OF PHIYY Bacteriophage phiYY was isolated
from sewage of Southwest Hospital, Chongqing, China, with _P. aeruginosa_ PAO38 as the host. phiYY forms clear plaques (2–4 mm in diameter) on double agar plates. Phage particles were very
sensitive to chloroform and could not form any plaques after chloroform treatment. The chloroform sensitivity of phiYY indicates a lipid membrane. The optimum multiplicity of infectivity
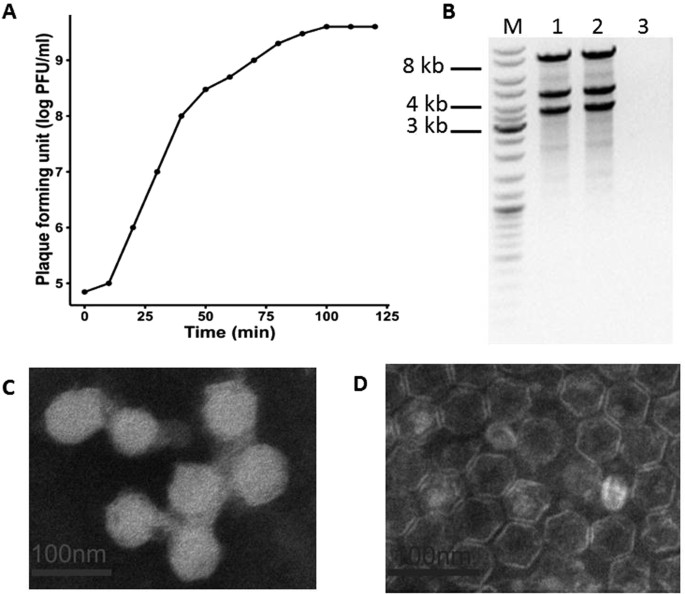
(MOI) of phiYY was observed as 0.1 (phage:host = 1:10). The one-step growth curve of phiYY showed that its latent period is approximately 10 min and that its burst period is approximately 50
min (Fig. 1A), which is considerably faster than that of phi6. However, the average burst size (10–14) was much lower than that of phi6 (125–400). Enzyme digestion showed that phiYY
contained three double-stranded RNA segments, which can be digested by RNase but not DNase (Fig. 1B). Transmission electron microscopy (TEM) revealed that the mature particle was tailless,
spherical, and approximately 50 nm in diameter (Fig. 1C). However, the chloroform-treated particles were icosahedral symmetric structures (Fig. 1D). IDENTIFICATION AND ORGANIZATION OF PHIYY
GENES The dsRNA genome of phiYY was reverse transcribed into complementary DNA (cDNA) fragments and sequenced by next-generation sequencing. The sizes of the segments were 3,004 (S), 3,862
(M), and 6,648 (L) bp, similar to the genome segments of phi6, which are 2948 (S), 4063 (M), and 6374 (L) bp. The GC-content was 57.46%, 58.91%, and 59.34% for phiYY S, M, and L,
respectively. A total of 20 potential open reading frames (ORFs) were predicted from the phiYY genome. (Fig. 2). Table 1 lists the putative functions of each gene. Among the 20 predicted
proteins of phiYY, 70% have putative functions. The majority of the phiYY proteins are structural proteins, such as the procapsid protein, host attachment protein, nucleocapsid shell
protein, muramidase, and RNA-dependent RNA polymerase (RdRP). Like most RNA viruses, phiYY likely packed RdRP into the virus during the infection cycle for later use to replicate phage RNA
upon host infection. Protein BLAST revealed that most structural proteins of phiYY are similar to those of other dsRNA bacteriophages. The _orf3_ gene, which encodes a polymerase4,18, showed
71% similarity to RdRP of phi13. The Orf5 protein was predicted to be an NTPase with 35% and 47% similarity to phi6 and phi13, respectively. NTPase is required for genomic packaging in
phi619. We also identified a muramidase, which showed 45% similarity to phi13, from segment S. Muramidase is a phage-encoded lysin that damages bacterial cell walls to release phage
particles during the last step of the phage infection cycle20,21. Given the emergence of multi-drug resistant bacteria, phage lysin has been suggested to be a potential antimicrobial
agent22. However, all the currently tested lysins were cloned from dsDNA phages. Thus, the efficiency of dsRNA phage-encoded muramidase against _P. aeruginosa_ should be tested in the near
future. Only 6 putative proteins were predicted with unknown functions. By contrast, most dsDNA phage genome contains numerous genes with unknown functions17,23; some potentially encode
virulent genes. Potential virulence is a safety concern for dsDNA phage therapy. However, dsRNA phage genomes are very concise and hypothetical proteins are very limited. Therefore, phiYY is
less likely to carry a toxin gene. This feature might be advantageous for dsRNA phage-based phage therapy. IDENTIFICATION OF PHIYY STRUCTURAL PROTEINS Structural proteins of phiYY were
analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The migration of the proteins was similar to that of other Cystoviridae (Fig. 3A). A total of 9 proteins
were detected with molecular weights of 10–80 kDa. To detect all the structural proteins, we performed SDS-PAGE for 30 min. We performed HPLC-MS analysis with the excised band containing the
mixture of all the structural proteins (Fig. 3B). All the structural proteins were detected. Interestingly, we also identified a hypothetical structural protein, Orf20, without similarity
to other phages. It is an additional structural protein absent in other dsRNA phages. Given that protein function cannot be determined by BLAST, further investigations are necessary to
characterize the function of Orf20. PHYLOGENETIC ANALYSIS Phylogenetic trees were generated by MEGA6 based on the nucleotide sequence alignment of Orf06 with the major capsid protein from
other dsRNA phages, phi6, phi8, phi12, phi13, phi2954, and phiNN (Fig. 4A). The nucleotide sequences of the phiYY L segment were also compared (Fig. 4B). The two generated phylogenetic trees
indicated that phiYY and phi13 are closely related. The _Pseudomonas_ phage phi13 was isolated from the leaves of the radish plant with _P. syringae_ as the host strain15. HOST RANGE To
characterize the host range of phiYY, we tested the phage against a panel of 233 isolated clinical strains and the type strain PAO1. The clinical strains were collected from seven hospitals
from four provinces, as described in the Materials and Methods section. The number of clinical strains from each hospital is indicated in Fig. 5B. To test the genetic diversity of our
collected strains, we performed _Enterobacteriaceae_ repetitive intergenic consensus polymerase chain reaction (ERIC-PCR)24. Supplementary Fig. S1 is a representative image of the ERIC-PCR
profiles from _P. aeruginosa_ strains. A total of 16 clusters were generated for the analyzed isolates, which were designated as Clusters 1 to 16. Figure 5A shows that the majority of our
collections can be assigned to Cluster 5 (98), Cluster 9 (36), Cluster 1 (35), and Cluster 4 (20). According to the ERIC-PCR study, our collected isolates exhibit high genetic diversity,
which can be used to assay the host range of phiYY. We used the dot plaque assay to determine the resistance/susceptibility of all the strains against phiYY. The formation of a clear plaque
in the bacterial lawn indicated susceptibility. Dot plaque assays were repeated to validate the results. In total, 233 clinical strains and PAO1 were tested. The host range was 42.3% (99 out
of 234). Supplementary Table 2 shows the detailed resistance or susceptibility map. phiYY was isolated from Southwest Hospital; hence, we collected 94 clinical strains from the same
hospital. The collected strains were classified into 11 different clusters by ERIC-PCR (Supplementary Table S3). 42.6% of these strains are sensitive to phiYY. By contrast, the majority of
the bacterial isolates from two hospitals in Sichuan Province were not lysed by phiYY (Fig. 5B). Additionally, phiYY did not infect PAO1. Overall, our _P. aeruginosa_ collections are
genetically diverse and phiYY infects 42.3% of these strains. _P. aeruginosa_ is an important Gram-negative opportunistic pathogen that causes serious infections in cystic fibrosis patients,
ICU patients, and other immune-compromised individuals25,26. Given that _P. aeruginosa_ is becoming resistant to most antibiotics, a common infection may be fatal. Therefore, the
feasibility of treating bacterial infections with bacteriophages has been extensively studied27,28,29,30,31. Several studies have reported the efficacy of phage therapy against _P.
aeruginosa_. The Phagoburn project was recently launched to systemically evaluate the safety and efficacy of phage therapy against _P. aeruginosa_ and _E. coli_ in burn patients32.
Interestingly, all the phages used in these studies are DNA phages. The effectiveness of dsRNA phage therapy has yet to be reported. dsRNA phages are possible alternative choices for phage
therapy. First, the dsRNA phage phiYY has a broad host range and can be used alone or as a component of a phage cocktail. Second, dsRNA phages have a higher mutation rate and host range
mutation rate33,34. The host range mutation frequency of the dsRNA bacteriophage phi6 is approximately 3 × 10−4. Therefore, the high host range mutation rate of dsRNA phage can be used to
expand or change the phage host range to better fit phage therapy. CONCLUSIONS This work presented a detailed analysis of the first dsRNA bacteriophage infecting _P. aeruginosa_. Our work
enhances the current understanding of RNA phage biology and provides an alternative choice for phage therapy. Given that phiYY infects 99 out of 233 clinical _P. aeruginosa_ strains, we are
interested in further expanding the dsRNA phage host range by evolution experiments and genetic engineering. We will also test the effectiveness of dsRNA phage therapy as compared with dsDNA
bacteriophages in animal models. MATERIALS AND METHODS BACTERIA AND BACTERIOPHAGE GROWTH CONDITIONS The _P. aeruginosa_ PAO1 strain was maintained in our lab. A total of 233 clinical
strains was collected from the Southwest Hospital (Chongqing, China), Daping Hospital (Chongqing, China), Xinqiao Hospital (Chongqing, China), West China Hospital (Sichuan, China), Sichuan
Provincial People’s Hospital (Sichuan, China), Xijin Hospital (Xi’an, China), and the Henan Provincial People’s Hospital (Henan, China). All of the bacteria were grown at 37 °C in LB medium.
phiYY was isolated from Southwest hospital sewage as previously described27. TRANSMISSION ELECTRON MICROSCOPY The morphology of the purified phage and the chloroform-treated phiYY was
observed by transmission electron microscopy as previously described25. Chloroform treatment was performed by vigorously shaking a mixture of 1 ml phiYY and 1 ml chloroform for 2 min. The
mixture was centrifuged at 13,000 × _g_ for 1 min. The phage particles were collected from the uppermost layer. ONE-STEP GROWTH CURVE To determine the one-step growth curve of phiYY, we used
the methods described by Lu _et al_.22. BACTERIOPHAGE RNA ISOLATION phiYY virions were purified as previously described. Briefly, phiYY was inoculated into the log phase host in LB medium
and cultured with aeration at 37 °C for 6 h. The phage lysate was centrifuged for 10 min at 10,000 × _g_ and passed through a 0.22-μm filter. The phages were further purified by PEG8000
precipitation. The phage dsRNA genome was isolated from the purified phage particles with the Tri Reagent extraction kit (Sigma-Aldrich). RNA integrity and size distribution were assessed on
a 1% (w/v) agarose gel and visualized with ethidium bromide. BACTERIOPHAGE GENOME SEQUENCING AND BIOINFORMATICS ANALYSIS Phage dsRNA genome was reverse-transcribed into cDNA with random
oligo primers (SuperScript® III First-Strand Synthesis System for RT-PCR, Invitrogen). Then, cDNA was sequenced by the semiconductor sequencer Ion Torrent Personal Genome Machine (PGM,
ThermoFisher). The data generated from the genomic library was approximately 12 M with an average read length of 223 bases. The read data was assembled by the de novo assembly algorithm
Newbler Version2.9 with default parameters. phiYY genes were predicted with RAST (http://rast.nmpdr.org/)35 and fgenesV
(http://linux1.softberry.com/berry.phtml?topic=virus&group=programs&subgroup=gfi ndv). The results were merged manually. DNA and protein sequences were scanned for homologs with
BLAST36. PROTEOMIC ANALYSIS OF PHAGE STRUCTURAL PROTEINS Proteomic analysis was performed as previously described23. Briefly, purified particles were denatured with heat and loaded onto a
15% (w/v) polyacrylamide gel. Proteins were stained with Coomassie Brilliant Blue R250 dye and washed with methanol–acetic acid–H2O. To identify phage structural proteins, we performed the
above mentioned experiment for 30 min. The protein band that included all the structural proteins was excised from the gel for HPLC-MS analysis. HPLC-MS data were processed by the Agilent
Spectrum Mill proteomics software to allocate each protein to the corresponding gene. ERIC-PCR TYPING Bacterial genomic DNA was extracted with TIANamp Genomic DNA Kit (DP304 Beijing, China).
PCR amplification was performed with the following primers24. ERIC1 (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC2 (5′-AAGTAAGTACTGGGGTGAGCG-3′). Each ERIC-PCR test was performed twice to ensure
the conformity of each fingerprint. The TIFF image was created from a photograph taken with the UV Gel Doc system (BIO-RAD, USA). The DNA banding patterns were entered into a database in
ImageLab 2.0 software (BIO-RAD, USA) to automatically obtain and analyze the images. The ERIC-PCR patterns were interpreted and compared as previously described24. Similarity analysis was
calculated with the Dice coefficient and the unweighted pair group average (UPGMA) for cluster analyses. When 80% or more similar bands were detected, these colonies were classified in the
same cluster. HOST RANGE ANALYSIS BY DOT PLAQUE ASSAY Phage sensitivity was determined by dot plaque assay. Briefly, 5 ml of molten 0.7% LB agar containing 100 μl of each test bacterial
culture was overlaid on 1.5% LB agar plates. Subsequently, 1 μl of phiYY (~109 PFU/ml) was spotted on the soft agar. Phage resistance/susceptibility was determined by the formation of clear
plaques after overnight culture at 37 °C. NUCLEOTIDE SEQUENCE ACCESSION NUMBER phiYY genomic sequence was submitted to GenBank. The accession numbers for segments L, M, and S are KX07420,
KX074202, and KX074203, respectively. ADDITIONAL INFORMATION HOW TO CITE THIS ARTICLE: Yang, Y. _et al_. Characterization of the first double-stranded RNA bacteriophage infecting
_Pseudomonas aeruginosa. Sci. Rep._ 6, 38795; doi: 10.1038/srep38795 (2016). PUBLISHER'S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and
institutional affiliations. REFERENCES * Manrique, P. et al. Healthy human gut phageome. Proceedings of the National Academy of Sciences of the United States of America, doi:
10.1073/pnas.1601060113 (2016). * Krishnamurthy, S. R., Janowski, A. B., Zhao, G., Barouch, D. & Wang, D. Hyperexpansion of RNA Bacteriophage Diversity. PLoS biology 14, e1002409, doi:
10.1371/journal.pbio.1002409 (2016). Article CAS PubMed PubMed Central Google Scholar * Vidaver, A. K., Koski, R. K. & Van Etten, J. L. Bacteriophage phi6: a Lipid-Containing Virus
of Pseudomonas phaseolicola. J Virol 11, 799–805 (1973). CAS PubMed PubMed Central Google Scholar * Sen, A. et al. Initial location of the RNA-dependent RNA polymerase in the
bacteriophage Phi6 procapsid determined by cryo-electron microscopy. The Journal of biological chemistry 283, 12227–12231, doi: 10.1074/jbc.M710508200 (2008). Article CAS PubMed PubMed
Central Google Scholar * Poranen, M. M., Daugelavicius, R., Ojala, P. M., Hess, M. W. & Bamford, D. H. A novel virus-host cell membrane interaction. Membrane voltage-dependent
endocytic-like entry of bacteriophage straight phi6 nucleocapsid. The Journal of cell biology 147, 671–682 (1999). Article CAS PubMed PubMed Central Google Scholar * de Haas, F.,
Paatero, A. O., Mindich, L., Bamford, D. H. & Fuller, S. D. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage phi6. Journal of molecular
biology 294, 357–372, doi: 10.1006/jmbi.1999.3260 (1999). Article CAS PubMed Google Scholar * Sinclair, J. F., Tzagoloff, A., Levine, D. & Mindich, L. Proteins of bacteriophage phi6.
J Virol 16, 685–695 (1975). CAS PubMed PubMed Central Google Scholar * Mindich, L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6.
Microbiology and molecular biology reviews: MMBR 63, 149–160 (1999). CAS PubMed PubMed Central Google Scholar * Katz, A. et al. Bacteriophage phi6–structure investigated by fluorescence
Stokes shift spectroscopy. Photochem Photobiol 88, 304–310, doi: 10.1111/j.1751-1097.2011.01051.x (2012). Article CAS PubMed Google Scholar * Qiao, X. Y., Sun, Y., Qiao, J., Di Sanzo, F.
& Mindich, L. Characterization of Phi 2954, a newly isolated bacteriophage containing three dsRNA genomic segments. BMC Microbiol. 10, doi: 10.1186/1471-2180-10-55 (2010). Article
PubMed PubMed Central Google Scholar * Mantynen, S. et al. New enveloped dsRNA phage from freshwater habitat. Journal of General Virology 96, 1180–1189, doi: 10.1099/vir.0.000063 (2015).
Article CAS PubMed Google Scholar * Vidaver, A. K., Koski, R. K. & Van Etten, J. L. Bacteriophage phi6: a Lipid-Containing Virus of Pseudomonas phaseolicola. J. Virol. 11, 799–805
(1973). CAS PubMed PubMed Central Google Scholar * Gottlieb, P., Wei, H., Potgieter, C. & Toporovsky, I. Characterization of variant phi12, a bacteriophage related to variant phi6:
Nucleotide sequence of the small and middle double-stranded RNA. Virology 293, 118–124, doi: 10.1006/viro.2001.1288 (2002). Article CAS PubMed Google Scholar * Hoogstraten, D. et al.
Characterization of phi8, a bacteriophage containing three double-stranded RNA genomic segments and distantly related to Phi6. Virology 272, 218–224, doi: 10.1006/viro.2000.0374 (2000).
Article CAS PubMed Google Scholar * Qiao, X., Qiao, J., Onodera, S. & Mindich, L. Characterization of phi 13, a bacteriophage related to phi 6 and containing three dsRNA genomic
segments. Virology 275, 218–224, doi: 10.1006/viro.2000.0501 (2000). Article CAS PubMed Google Scholar * Le, S. et al. Mapping the tail fiber as the receptor binding protein responsible
for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS One 8, e68562, doi: 10.1371/journal.pone.0068562 (2013). Article CAS ADS PubMed PubMed
Central Google Scholar * Shen, M. et al. Characterization and Comparative Genomic Analyses of Pseudomonas aeruginosa Phage PaoP5: New Members Assigned to PAK_P1-like Viruses. Sci Rep 6,
34067, doi: 10.1038/srep34067 (2016). Article CAS ADS PubMed PubMed Central Google Scholar * Butcher, S. J., Makeyev, E. V., Grimes, J. M., Stuart, D. I. & Bamford, D. H.
Crystallization and preliminary X-ray crystallographic studies on the bacteriophage phi6 RNA-dependent RNA polymerase. Acta crystallographica. Section D, Biological crystallography 56,
1473–1475 (2000). Article CAS PubMed Google Scholar * Nemecek, D., Heymann, J. B., Qiao, J., Mindich, L. & Steven, A. C. Cryo-electron tomography of bacteriophage phi6 procapsids
shows random occupancy of the binding sites for RNA polymerase and packaging NTPase. Journal of structural biology 171, 389–396, doi: 10.1016/j.jsb.2010.06.005 (2010). Article CAS PubMed
PubMed Central Google Scholar * Sharma, M., Kumar, D. & Poluri, K. M. Elucidating the pH-Dependent Structural Transition of T7 Bacteriophage Endolysin. Biochemistry 55, 4614–4625, doi:
10.1021/acs.biochem.6b00240 (2016). Article CAS PubMed Google Scholar * Kutyshenko, V. P. et al. Structure and dynamics of the retro-form of the bacteriophage T5 endolysin. Biochimica
et biophysica acta 1864, 1281–1291, doi: 10.1016/j.bbapap.2016.06.018 (2016). Article CAS PubMed Google Scholar * Na, H., Kong, M. & Ryu, S. Characterization of LysPBC4, a novel
Bacillus cereus-specific endolysin of bacteriophage PBC4. FEMS microbiology letters 363, doi: 10.1093/femsle/fnw092 (2016). * Lu, S. et al. Genomic and proteomic analyses of the terminally
redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages. PLoS One 8, e62933, doi: 10.1371/journal.pone.0062933 (2013). Article CAS ADS PubMed
PubMed Central Google Scholar * Khosravi, A. D., Hoveizavi, H., Mohammadian, A., Farahani, A. & Jenabi, A. Genotyping of multidrug-resistant strains of Pseudomonas aeruginosa isolated
from burn and wound infections by ERIC-PCR. Acta Cir. Bras. 31, 206–211, doi: 10.1590/s0102-865020160030000009 (2016). Article PubMed Google Scholar * Soothill, J. Use of bacteriophages
in the treatment of Pseudomonas aeruginosa infections. Expert review of anti-infective therapy 11, 909–915, doi: 10.1586/14787210.2013.826990 (2013). Article CAS PubMed Google Scholar *
Saussereau, E. & Debarbieux, L. Bacteriophages in the experimental treatment of Pseudomonas aeruginosa infections in mice. Advances in virus research 83, 123–141, doi:
10.1016/B978-0-12-394438-2.00004-9 (2012). Article CAS PubMed Google Scholar * Pires, D. P., Boas, D. V., Sillankorva, S. & Azeredo, J. Phage Therapy: a Step Forward in the Treatment
of Pseudomonas aeruginosa Infections. J. Virol. 89, 7449–7456, doi: 10.1128/jvi.00385-15 (2015). Article CAS PubMed PubMed Central Google Scholar * Olszak, T. et al. _In vitro_ and _in
vivo_ antibacterial activity of environmental bacteriophages against Pseudomonas aeruginosa strains from cystic fibrosis patients. Applied microbiology and biotechnology 99, 6021–6033, doi:
10.1007/s00253-015-6492-6 (2015). Article CAS PubMed PubMed Central Google Scholar * Essoh, C. et al. Investigation of a Large Collection of Pseudomonas aeruginosa Bacteriophages
Collected from a Single Environmental Source in Abidjan, Cote d’Ivoire. PLoS One 10, e0130548, doi: 10.1371/journal.pone.0130548 (2015). Article CAS PubMed PubMed Central Google Scholar
* Danis-Wlodarczyk, K. et al. Correction: Characterization of the Newly Isolated Lytic Bacteriophages KTN6 and KT28 and Their Efficacy against Pseudomonas aeruginosa Biofilm. PLoS One 10,
e0137015, doi: 10.1371/journal.pone.0137015 (2015). Article CAS PubMed PubMed Central Google Scholar * Beeton, M. L., Alves, D. R., Enright, M. C. & Jenkins, A. T. A. Assessing
phage therapy against Pseudomonas aeruginosa using a Galleria mellonella infection model. Int. J. Antimicrob. Agents 46, 196–200, doi: 10.1016/j.ijantimicag.2015.04.005 (2015). Article CAS
PubMed Google Scholar * Resch, G. et al. Update on PHAGOBURN, the First European Clinical Trial of Phage Therapy. Abstracts of the General Meeting of the American Society for
Microbiology 114, 1277 (2014). Google Scholar * Ferris, M. T., Joyce, P. & Burch, C. L. High frequency of mutations that expand the host range of an RNA virus. Genetics 176, 1013–1022,
doi: 10.1534/genetics.106.064634 (2007). Article CAS PubMed PubMed Central Google Scholar * Ford, B. E. et al. Frequency and Fitness Consequences of Bacteriophage Phi 6 Host Range
Mutations. PLoS One 9, doi: 10.1371/journal.pone.0113078 (2014). * Brettin, T. et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation
pipelines and annotating batches of genomes. Sci Rep 5, 8365, doi: 10.1038/srep08365 (2015). Article CAS PubMed PubMed Central Google Scholar * Singh, H. & Raghava, G. P.
BLAST-based structural annotation of protein residues using Protein Data Bank. Biology direct 11, 4, doi: 10.1186/s13062-016-0106-9 (2016). Article CAS PubMed PubMed Central Google
Scholar Download references ACKNOWLEDGEMENTS This work was supported by the National Natural Science Foundation of China (NSFC, 31501004 to S.Le). We thank Prof. Tong Yigang for the
sequencing of phage phiYY genome. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Microbiology, Third Military Medical University, Chongqing, 400038, China Yuhui Yang, Shuguang
Lu, Wei Shen, Xia Zhao, Mengyu Shen, Yinling Tan, Gang Li, Ming Li, Jing Wang, Fuquan Hu & Shuai Le Authors * Yuhui Yang View author publications You can also search for this author
inPubMed Google Scholar * Shuguang Lu View author publications You can also search for this author inPubMed Google Scholar * Wei Shen View author publications You can also search for this
author inPubMed Google Scholar * Xia Zhao View author publications You can also search for this author inPubMed Google Scholar * Mengyu Shen View author publications You can also search for
this author inPubMed Google Scholar * Yinling Tan View author publications You can also search for this author inPubMed Google Scholar * Gang Li View author publications You can also search
for this author inPubMed Google Scholar * Ming Li View author publications You can also search for this author inPubMed Google Scholar * Jing Wang View author publications You can also
search for this author inPubMed Google Scholar * Fuquan Hu View author publications You can also search for this author inPubMed Google Scholar * Shuai Le View author publications You can
also search for this author inPubMed Google Scholar CONTRIBUTIONS S.L. and F.H. conceived the study. Y.Y., S.L., W.S., Z.X. performed the experiments. M.S., Y.T., G.L., M.L., J.W. analyzed
the sequence data. S.L. wrote the paper. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY
INFORMATION RIGHTS AND PERMISSIONS This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included
in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain
permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Yang, Y., Lu, S., Shen, W. _et al._ Characterization of the first double-stranded RNA bacteriophage infecting _Pseudomonas aeruginosa_. _Sci Rep_ 6, 38795 (2016).
https://doi.org/10.1038/srep38795 Download citation * Received: 08 September 2016 * Accepted: 14 November 2016 * Published: 09 December 2016 * DOI: https://doi.org/10.1038/srep38795 SHARE
THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative
