Isolation of a new antibacterial peptide achromosin from streptomyces achromogenes subsp. Achromogenes based on genome mining
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

Lasso peptides are a class of ribosomally biosynthesized and post-translationally modified peptides with a common motif of knot structure in the molecule.1 The amino group of the N-terminal
amino acid forms a peptide bond with side chain carboxyl group of Asp or Glu in the eighth or the ninth position from the N-terminus, resulting in formation of a macrolactam ring. The
macrolactam ring looks like a loop of a ‘lasso’ with a tail of the C-terminal linear peptide that normally locates through the ring. Regarding lasso peptides, a wide variety of biological
activities such as anti-HIV,2 antimycobacterial,3 endothelin type B receptor antagonist4 and prolyl endopeptidase inhibition5 were reported. In addition, lasso peptides normally show a
stable property against proteolytic, thermal and chemical degradation, which makes lasso peptides attractive in terms of practical application as pharmaceutical reagents. Lasso peptides
derived from actinobacteria have been classified into three main classes on the basis of their _N_-terminal residues and the number of disulfide bridges.1 The class I lasso peptides include
siamycins I and II,2 aborycin6 and sviceucin,7 which have an internal peptide linkage between β-carboxyl group residue of Asp9 (ninth amino acid residue from the N-terminus) and the amino
residue of Cys1. These peptides commonly have additional two disulfide bridges between Cys1 and Cys13, and Cys7 and Cys19. The class II lasso peptides include anantin,8 lariatins,3
propeptin,5 RES-701-1,4 SRO15-20059 and sungsanpin.10 These peptides have an internal peptide linkage between β-carboxyl residue of Asp8 or Asp9 and the amino residue of Gly1 without any
disulfide bonds. The class III lasso peptide includes only one peptide named BI-32169.11 The peptide BI-32169 has an internal peptide linkage between β-carboxyl residue of Asp9 and the amino
residue of Gly1 with one disulfide bond between Cys6 and Cys19. The lasso peptide microcin J25 was isolated from _Escherichia coli_, which is regarded as the archetype of lasso peptides.12
Its biosynthetic gene cluster consists of four genes including a precursor peptide-coding gene: gene A (_mcjA_), two maturation enzymes including gene B (_mcjB,_ cleavage of leader peptide)
and gene C (_mcjC_, formation of macrolactam ring) and an ATP-binding cassette transporter-coding gene: gene D (_mcjD_).13 The protein McjC was reported to form the macrolactam ring, and the
function of the protein McjB was assigned to cleave off the leader peptide from the precursor peptide by in _vitro_ experiments.14 Normally lasso peptide biosynthetic genes in
proteobacteria have a corresponding set of the genes, although the transporter gene is optional.1 In actinobacteria, lasso peptide biosynthetic genes consist of a similar gene set, except
that a maturation enzyme gene B has split-B genes (gene B1 and gene B2).1, 15 By genome mining, biosynthetic genes of a lasso peptide sviceucin were found on the genome of _Streptomyces
sviceus_, and the lasso peptide was isolated and structure-determined by heterologous expression.7 The lasso peptide SRO15-2005 was identified by matrix-assisted laser
desorption/ionization-time-of-flight tandem mass spectrometry (MALDI-TOF-MS/MS) from the extract of _Streptomyces roseosporus_, based on genome sequence data.9 On the basis of genome mining,
a new lasso peptide chaxapeptin was also isolated as a lung cancer invasion inhibitor from _Streptomyces leeuwenhoekii._16 These results prompted us to find a new lasso peptide from
streptomycetes using genome sequence data. By genome search approach, we found new lasso peptide biosynthetic genes on the genome sequence of _Streptomyces achromogenes_ subsp.
_achromogenes_.17 The new antibacterial peptide was isolated by chromatographic separation from the culture of _S. achromogenes subsp. achromogenes_. Here, we describe isolation and
structure determination of a new antibacterial peptide named achromosin. In the genome sequence of _Streptomyces achromogenes_ subsp. _achromogenes_,17 lasso peptide modification
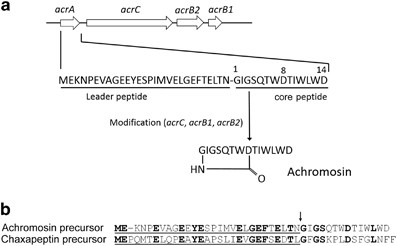
enzyme-coding genes (gene C named _acrC_: WP_063755122.1, _acrB2_: WP_037654156.1, _acrB1_: WP_037654159.1, shown in Figure 1a and Supplementary Table S1) were found by blastp similarity
search. As the lasso precursor peptide-coding gene was not annotated, we searched for the lasso precursor peptide-coding gene in the close region to the modification enzyme-coding genes.
Upstream of the gene _acrC_ (WP_063755122.1), a new putative precursor peptide-coding gene for new peptide named achromosin (126 base pairs, 42 amino acids, Figure 1b) similar to
chaxapeptin16 was found from position 72 827 to 72 952 bp in the genome sequence (GenBank accession number: NZ_JODT01000002.1). On the upstream of 9 residues of the precursor peptide-coding
region (72827-72952), Shine–Dalgarno sequence (AGGAGGA) was present. As shown in Figure 1b, the expected peptide achromosin was deduced to have the amino acid sequence of GIGSQTWDTIWLWD
(monoisotopic molecular weight: 1676.7 Da), after cleaving off the leader peptide at the same position after the conserved motif ‘GEFXEXTX’ as the biosynthesis of chaxapeptin16 (arrow in
Figure 1b). The expected monoisotopic molecular weight of achromosin was calculated to be 1658.7 Da considering the loss of 18 Da, resulting in macrolactam formation of lasso peptide
biosynthesis. The preliminary chemical investigation of _S. achromogenes_ subsp. _achromogenes_ NBRC12735T indicated that the expected peptide was present in the methanol extract of aerial
hyphae and spore cells by high-performance liquid chromatography (HPLC) and electrospray ionization mass spectrometry (ESI-MS). Thus, cultivation of _S. achromogenes_ subsp. _achromogenes_
was performed in a large scale to obtain enough amount of the peptide for structure determination. After 7 days of cultivation, cells of spore and aerial hyphae were harvested by a steel
spatula. The cells were extracted with double volume of methanol (MeOH), followed by centrifugation. After condensation using rotary evaporation, the extract was subjected to open-column
chromatography using hydrophobic resin (CHP-20P), eluted with 10%, 60% and 100% MeOH. The expected peptide achromosin was detected in 100% MeOH fraction by HPLC (Supplementary Figure S1) and
ESI-MS analysis (Supplementary Figure S2). The ESI-MS analysis of the peptide gave an ion peak at _m/z_ 1659.7 for [M+H]+. The 100% MeOH fraction was repeatedly subjected to HPLC
purification to give pure achromosin. The molecular formula of achromosin was established to be C79H106N18O22 by accurate mass analysis using the ESI Fourier-transform ion cyclotron
resonance mass spectrometry, as [M+2H]2+ was observed at _m/z_ 830.3941 corresponding to C79H108N18O22 whose calculated value was 830.3937. The amino acid composition analysis was performed
on achromosin following the reported method.18 The amino acid content analysis on achromosin afforded the relative molar ratios of the constituent amino acids (2 moles each of Asp/Asn, Gly,
Ile and Thr, and 1 mole each of Glu/Gln, Leu and Ser), as shown in Supplementary Figure S3. Nuclear magnetic resonance analysis using dimethyl sulfoxide-_d_6 as a solvent was not possible
due to ambiguous broad peaks in the nuclear magnetic resonance spectrum. To obtain peptide sequence, MALDI-TOF-MS/MS analysis on achromosin was accomplished. As a result, the product ions
from achromosin at _m/z_ 1659 were of _b_-series peptides, _b_8-_b_13 (Figure 2a and Supplementary Table S2), which indicated that the sequence of TIWLWD was the C-terminus tail sequence.
Macrolactam ring structure was reported not to give fragment ions,9 thus we proposed the structure of achromosin to be shown in Figure 2a, based on the amino acid sequence of precursor
peptide gene. To confirm the amino acid sequence in the macrolactam ring, C-terminal peptide bonds of tryptophans were cleaved by BNPS-skatole. After BNPS-skatole reaction, the cleaved
achromosin (BNPS-achromosin) was purified by HPLC separation. ESI-TOF-MS analysis on BNPS-achromosin gave an ion peak at _m/z_ 1291.5 for [M+H]+ (Supplementary Figure S4). The molecular
formula of BNPS-achromosin was clarified to be C58H78N14O20 by the accurate mass analysis. That is, [M+2H]2+ was observed at _m/z_ 646.2832 corresponding to C58H80N14O20 whose calculated
value was 646.2831. By the reaction of BNPS-skatole the Trp residue in a peptide is oxidized and transformed to 3-oxindole with a spirolactone, which increases the molecular weight due to
the addition of two oxygens by 32 Da. As shown in Figure 2b, the MALDI-TOF-MS/MS of the cleaved achromosin gave the sequence of the peptide with one N-terminus and two C-terminal ends
(Supplementary Table S3). The product ions of _b1_, _b2_ and _b3_ supported the sequence of DTIW* and _b4_ ion especially indicated that Trp at C-terminus was oxidized (indicated with
asterisk, Figure 2b). The product ions of _y2_ to _y7_ supported the sequence of GIGSQTW* (Figure 2b). Above all, the structure of achromosin was proposed to be a peptide with the sequence
of GIGSQTWDTIWLWD having one macrolactam ring which was formed by peptide bond between amino residue of Gly1 and β-carboxyl residue of Asp8 (Figure 2a). The structure of achromosin did not
include any disulfide bridge in the molecule, which classified achromosin into class II lasso peptide. The antimicrobial activity of achromosin was tested using a paper disk agar-diffusion
assay against microorganisms (bacterial strains including _E. coli_, _Pseudomonas aeruginosa_, _Serratia marcescens_, _Bacillus subtilis_, _Staphylococcus aureus_, _Micrococcus luteus_ and
_Streptomyces antibioticus_; Yeast strains including _Saccharomyces cerevisiae_, _Schizosaccharomyces pombe_, _Kloeckera apiculata_; fungi strains including _Aspergillus niger_, _Aspergillus
oryzae_ and _Mucor hiemalis_). At the dosage of 10 μg per disk, achromosin showed an inhibitory zone of 11 mm diameter against _M. luteus_ (Supplementary Figure S5). On the other hand,
achromosin did not show any inhibitory activity against the other testing microorganisms at the same dosage. Biosynthetic gene clusters of lasso peptides of actinobacteria have been
identified for lasso peptides including lariatin,19 SRO15-2005,9 lassomycin,20 sviceucin,7 chaxapeptin16 and streptomonomicin.21 The biosynthetic gene cluster of chaxapeptin consisted of
four genes including _cptA_, _cptC_, _cptB1_ and _cptB2_.16 Interestingly, the gene cluster of chaxapeptin lacked of transporter gene that often exists in the lasso peptide biosynthetic gene
cluster. The gene _cptA_ encoded chaxapeptin precursor peptide, and the three genes including _cptC_, _cptB1_ and _cptB2_ were proposed to be involved in macrolactam formation and leader
peptide cleavage. The amino acid sequence of precursor peptide gene _acrA_ which was found on the genome of _S. achromogenes_ subsp. _achromogenes_17 showed high similarity with that of
_cptA_ (46% identity, 68% positive matches). By reference to chaxapeptin biosynthetic genes, we assigned the biosynthetic gene cluster for achromosin, which have four genes, _acrA_ (annoted
in this study, 42 aa), _acrC_ (WP_063755122.1, 616 aa), _acrB2_ (WP_037654156.1, 150 aa) and _acrB1_ (WP_037654159.1, 95 aa) in this order with all the same direction (Figure 1a).
Interestingly, there was no transport protein-coding genes near the gene cluster. The lack of transport gene was also reported in the chaxapeptin gene cluster.16 On the basis of the
similarity of each gene, we proposed the functions of the genes as shown in Figure 1a. The gene _acrA_ encoded the precursor of achromosin and the genes including _acrC_, _acrB1_ and _acrB2_
were proposed to be modification enzymes to give the mature lasso peptide. The gene _acrC_ encoded putative asparagine synthase possibly responsible for formation of the Gly1–Glu8 amide
bond, which showed high similarity to _cptC_ by using a BLAST homology search (37% identity, 51% positive matches). The amino acid sequence of _acrB2_ showed high similarity to that of
_cptB2_ by using a BLAST homology search (55% identity, 69% positive matches) and the amino acid sequence of _acrB1_ showed high similarity to that of _cptB1_ by using a BLAST homology
search (40% identity, 54% positive matches). Above all, the biosynthetic genes of achromosin showed the similarity to those of chaxapeptin. So far, no similar peptide has been found by the
blastp search, which indicates the novelty of achromosin. As shown in Figure 1b, the amino acid sequence of core peptide is different even from that of chaxapeptin, the closest lasso
peptide. The lasso peptide in class II were reported to have a wide variety of biological activities such as antimycobacterial,3 endothelin type B receptor antagonist4 and prolyl
endopeptidase inhibition5. In this paper, the antimicrobial activity of achromosin was tested. Further bioactivity tests may lead to the discovery of additional activities of achromosin. In
addition, the biosynthetic genes of achromosin were identified from the genome of _S. achromogenes_ subsp. _achromogenes,_ which will lead to genetic engineering using the gene cluster to
create mutated lasso peptide based on achromosin by heterologous expression. The modified peptides with more potent antibacterial activity may be produced by altering the amino acid sequence
of achromosin by further genetic engineering experiments. ACCESSION CODES ACCESSIONS GENBANK/EMBL/DDBJ * NZ_JODT01000002.1 REFERENCES * Li, Y., Zirah, S. & Rebuffat, S. _Lasso Peptides:
Bacterial Strategies to Make and Maintain Bioactive Entangled Scaffolds_, Springer, (2015). Book Google Scholar * Detlefsen, D. J. _et al_. Siamycins I and II, new anti-HIV-1 peptides:
II. Sequence analysis and structure determination of siamycin I. _J. Antibiot._ 48, 1515–1517 (1995). Article CAS Google Scholar * Iwatsuki, M. _et al_. Lariatins, antimycobacterial
peptides produced by _Rhodococcus_ sp. K01-B0171, have a lasso structure. _J. Am. Chem. Soc._ 128, 7486–7491 (2006). Article CAS Google Scholar * Morishita, Y. _et al_. RES-701-1, a novel
and selective endothelin type B receptor antagonist produced by _Streptomyces_ sp. RE-701. I. Characterization of producing strain, fermentation, isolation, physico-chemical and biological
properties. _J. Antibiot._ 47, 269–275 (1994). Article CAS Google Scholar * Kimura, K. _et al_. Propeptin, a new inhibitor of prolyl endopeptidase produced by _Microbispora_. I.
Fermentation, isolation and biological properties. _J. Antibiot._ 50, 373–378 (1997). Article CAS Google Scholar * Potterat, O. _et al_. Tricyclic 21-peptide antibiotic isolated from
_Streptomyces griseoflavus_. _Liebigs. Ann._ 1994, 741–743 (1994). Article Google Scholar * Li, Y. _et al_. Characterization of sviceucin from _Streptomyces_ provides insight into enzyme
exchangeability and disulfide bond formation in lasso peptides. _ACS. Chem. Biol._ 10, 2641–2649 (2015). Article CAS Google Scholar * Weber, W. _et al_. Anantin-a peptide antagonist of
the atrial natriuretic factor (ANF). I. Producing organism, fermentation, isolation and biological activity. _J. Antibiot._ 44, 164–171 (1991). Article CAS Google Scholar * Kersten, R. D.
_et al_. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. _Nat. Chem. Biol._ 7, 794–802 (2011). Article CAS Google Scholar * Um, S. _et al_.
Sungsanpin, a lasso peptide from a deep-sea streptomycete. _J. Nat. Prod._ 76, 873–879 (2013). Article CAS Google Scholar * Potterat, O. _et al_. BI-32169, a bicyclic 19-peptide with
strong glucagon receptor antagonist activity from _Streptomyces_ sp. _J. Nat. Prod._ 67, 1528–1531 (2004). Article CAS Google Scholar * Salomon, R. A. & Farias, R. N. Microcin 25, a
novel antimicrobial peptide produced by _Escherichia coli_. _J. Bacteriol._ 174, 7428–7435 (1992). Article CAS Google Scholar * Solbiati, J. O. _et al_. Genetic analysis of plasmid
determinants for microcin J25 production and immunity. _J. Bacteriol._ 178, 3661–3663 (1996). Article CAS Google Scholar * Yan, K. P. _et al_. Dissecting the maturation steps of the lasso
peptide microcin J25 _in vitro_. _Chembiochem._ 13, 1046–1052 (2012). Article CAS Google Scholar * Burkhart, B. J. _et al_. A prevalent peptide-binding domain guides ribosomal natural
product biosynthesis. _Nat. Chem. Biol._ 11, 564–570 (2015). Article CAS Google Scholar * Elsayed, S. S. _et al_. Chaxapeptin, a lasso peptide from extremotolerant _Streptomyces
leeuwenhoekii_ strain C58 from the hyperarid atacama desert. _J. Org. Chem._ 80, 10252–10260 (2015). Article CAS Google Scholar * Ju, K. S. _et al_. Discovery of phosphonic acid natural
products by mining the genomes of 10,000 actinomycetes. _Proc. Natl Acad. Sci. USA_ 112, 12175–12180 (2015). Article CAS Google Scholar * Heinrikson, R. L. & Meredith, S. C. Amino
acid analysis by reverse-phase high-performance liquid chromatography: precolumn derivatization with phenylisothiocyanate. _Anal. Biochem._ 136, 65–74 (1984). Article CAS Google Scholar *
Inokoshi, J. _et al_. Molecular cloning of the gene cluster for lariatin biosynthesis of _Rhodococcus jostii_ K01-B0171. _Appl. Microbiol. Biotechnol._ 95, 451–460 (2012). Article CAS
Google Scholar * Gavrish, E. _et al_. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. _Chem. Biol._
21, 509–518 (2014). Article CAS Google Scholar * Metelev, M. _et al_. Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso peptide from an understudied
halophilic actinomycete. _Chem. Biol._ 22, 241–250 (2015). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This study was supported by the Japan Society for the Promotion
of Science by Grants-in-aids (grant number 25350964). We thank Ms Tomoko Satoh (Bruker Daltonics) for her technical assistance in the MS analysis. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS
* Graduate School of Integrated Science and Technology, Shizuoka University, Shizuoka, Japan Issara Kaweewan & Shinya Kodani * Food Research Institute, National Agriculture and Food
Research Organization (NARO), Ibaraki, Japan Mayumi Ohnishi-Kameyama * College of Agriculture, Academic Institute, Shizuoka University, Shizuoka, Japan Shinya Kodani * Graduate School of
Science and Technology, Shizuoka University, Shizuoka, Japan Shinya Kodani Authors * Issara Kaweewan View author publications You can also search for this author inPubMed Google Scholar *
Mayumi Ohnishi-Kameyama View author publications You can also search for this author inPubMed Google Scholar * Shinya Kodani View author publications You can also search for this author
inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Shinya Kodani. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION
Supplementary Information accompanies the paper on The Journal of Antibiotics website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (DOCX 695 KB) RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kaweewan, I., Ohnishi-Kameyama, M. & Kodani, S. Isolation of a new antibacterial peptide achromosin from _Streptomyces achromogenes_
subsp. _achromogenes_ based on genome mining. _J Antibiot_ 70, 208–211 (2017). https://doi.org/10.1038/ja.2016.108 Download citation * Received: 15 May 2016 * Revised: 03 July 2016 *
Accepted: 30 July 2016 * Published: 07 September 2016 * Issue Date: February 2017 * DOI: https://doi.org/10.1038/ja.2016.108 SHARE THIS ARTICLE Anyone you share the following link with will
be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative
