Opantimycin a, a new metabolite isolated from streptomyces sp. Rk88-1355
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

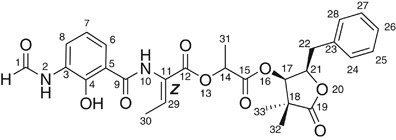
A new metabolite containing a γ-butyrolactone and 2-hydroxy-3-formylaminobenzoic acid moieties, opantimycin A (1) was isolated from a microbial metabolite fraction library generated from
_Streptomyces_ sp. RK88-1355 by search of an LC/MS-based spectral database named NPPlot: Natural Products Plot. The structure of 1 was determined based on extensive spectroscopic methods
including NMR, MS and MS/MS experiments. 1 showed moderate cytotoxicity against HL-60 cell lines and antimalarial activity against _Plasmodium falciparum_ 3D7. It was speculated that 1 might
be biosynthesized by a hybrid enzyme including non-ribosomal peptide synthetase and polyketide synthase, which were similar to neoantimycin biosynthetic machinery. Microbial metabolites
isolated from actinomycetes and fungi have unique and wide chemical diversity, and they are major sources in the discovery of novel drug candidates for various biological activities.1, 2
Hence they have played an important role in drug discovery and development of agrochemicals.3 They are also used as bioprobes, which are chemical tools to investigate biological functions in
chemical biology studies.4, 5 To discover and isolate such unique and important metabolites efficiently, we have constructed a microbial metabolite fraction library consisting of
semi-purified metabolites by basic chromatographic techniques such as HPLC and middle pressure liquid chromatography (MPLC) coupled with LC/MS-based spectral database named NPPlot.6, 7
NPPlot is a distribution map of metabolites, which are plotted as dots in two-dimensional area by retention time and _m_/_z_ value for _x_ and _y_ axes. Each metabolite has UV information of
maximum absorption values for _z_ axis in NPPlot, which allows us to find specific or distinctive metabolite groups easily. In the course of screening for structurally novel metabolites
from fraction libraries by NPPlot, we have discovered and isolated several new metabolites with interesting structures, such as verticilactam,8 spirotoamides9 and pyrrolizilactone.10 We have
recently reported new quinomycin derivatives, RK-1355A and B11 and a new neoantimycin analogue, unantimycin A12 from the fraction library of _Streptomyces_ sp. RK88-1355 by NPPlot search,
in which distinctive metabolites groups were screened by comparison of NPPlot generated from several _Streptomyces_ strains. Based on our continuing search for structurally unique
metabolites in the RK88-1355 fraction library, unidentified metabolites were found by NPPlot screening. They showed similar UV absorption pattern to those of neoantimycins.12, 13, 14
However, MWs of these compounds were around 550 Da, which was less than those of neoantimycins about 100 Da. One of these metabolites, a new compound (1), was isolated from the related
fraction by C18-HPLC. We report, herein, the isolation, structure and biological activities of 1 (Figure 1). Opantimycin A (1, 1.3 mg) was isolated as pale-yellow amorphous from the fraction
library generated from _Streptomyces_ sp. RK88-1355 by C18-HPLC. The molecular formula of 1 was determined to be C28H30N2O9 by high-resolution electrospray ionization mass spectrometry
(HRESIMS) (found: _m_/_z_ 539.2026 [M+H]+, calculated for C28H31N2O9, 539.2030). The IR spectrum implied the presence of hydroxyl (3330 cm−1), carbonyl (1756 and 1729 cm−1) and amide (1660
and 1525 cm−1) groups (Table 1). The 1H NMR spectrum suggested the presence of a benzene ring (_δ_H 7.21 of 2H, 7.28 of 2H and 7.23) and 1,2,3-trisubstituted benzene (_δ_H 6.90 dd [_J_=8.1,
8.1], 7.27 m and 8.54d [_J_=8.1]) (Table 2). It also showed a doublet signal at 8.48 ppm (_J_=1.2) and two exchangeable signals as broad peaks at 7.64 and 7.90 ppm, which were confirmed by
the addition of D2O and observation of disappearance of the signals in 1H NMR spectrum (Supplementary Figures S1 and S2). These observations suggested that 1 had a related structure to
neoantimycins with a benzene ring and a 2-hydroxy-3-formylaminobenzoic acid, which were representative functional groups for neoantimycins. This was also supported by unusual low-fielded
chemical shift value of H-8 at 8.54 ppm and a related UV absorption spectrum with those of neoantimycins such as SW-163A.12, 13 The 1H NMR spectrum also showed four of methyl signals
including two of singlet signals at 1.16 and 1.30 ppm and two of doublet signals at 1.60 (_J_=6.9) and 1.88 ppm (_J_=6.9), which were supposed to be attached on an sp2 carbon. The 13C NMR
spectrum showed 26 signals, two of which were observed as a double intensity supporting the presence of a benzene ring (Supplementary Figure S3). It also contained four carbonyl carbon
signals at 163.6, 168.7, 169.9 and 179.2 ppm, which were confirmed by 13C DEPT experiment (Supplementary Figure S4). The planar structure was investigated by the detailed interpretation of
2D NMR spectra including HSQC, DQF-COSY, HSQC-TOCSY, HMBC and phase-sensitive NOESY (Figure 2 and Supplementary Figures S5–S9). The connections between proton and carbon were confirmed by
the correlations observed in the HSQC spectrum. The proton spin networks between H-1 and NH-2, H-6 to H-8, H-14 and Me-31, H-17 to H-22, H-24 to H-28, and H-29 and H-30 were confirmed by the
correlations observed in DQF-COSY and HSQC-TOCSY (Figure 2a). A 2-hydroxy-3-formylaminobenzoic acid moiety was confirmed by the HMBC correlations from H-1 to C-3, H-6 to C-4 and C-9, H-7 to
C-3 and C-5, and H-8 to C-3 and C-4. A benzene moiety was confirmed by an HMBC correlation from H-25 and 27 to C-23 and attached to C-21 through a methylene carbon of C-22 assigned by HMBC
correlations from H-22 to C-23 and C-24 and 28. A partial structure from C-30 to C-22 were constructed by combination of consideration of 13C NMR chemical shift values of oxygenated carbons
at C-14 and C-17 and HMBC correlations from H-30 to C-11, H-29 to C-11 and C-12, H-14 to C-12, H-31 to C-15, H-17 to C-15 and C-19, and Me-32 and 33 correlating each other, to C-17, C-18 and
C-19. A γ-butyrolactone was constructed by the consideration of 13C NMR chemical shift value of C-19 at 179.2 ppm, C-21 at 80.3 ppm, 1H NMR chemical shift value of H-21 at 4.76 ppm (ddd,
_J_=9.1, 4.6, 3.4 Hz) and the index of hydrogen deficiency of 15. The connection between C-9 and N-10 was constructed by the NOESY correlation between H-6 and NH-10. C-11 was considered
connecting to N-10 by the 13C NMR chemical shift value of 124.9 ppm although no HMBC correlation was observed from NH-10 to C-11. To confirm the connection, MS/MS experiment was carried out,
and it showed fragment ions at _m_/_z_ 375, 247 and 164 in ESI-positive mode (Figure 2c). These observations supported the connection between NH-10 and C-11 and also C-9 and NH-10, and the
planar structure of 1 was confirmed. The geometry at Δ11 was deduced as _Z_-configuration by the comparison of 13C NMR chemical shift value of Me-30 at 15.3 ppm with those reported for
phomalide and isophomalide,15 which had identical dehydrobutyrine units of _E_- and _Z_-configuration (14.4 and 15.3 ppm in chloroform-_d_ for _E_- and _Z_-configuration) with that of 1. It
was also supported by the NOESY correlation between H-29 and Me-31 (Figure 2a). The relative stereochemistry on the γ-butyrolactone was assigned to have _cis_ configuration at C-17 and C-21
by the NOESY correlation between H-21 to both of H-17 and Me-33 (Figure 2b). As the result, the structure of 1 was determined as shown in Figure 1, designated as opantimycin A.
Cytotoxicities against human cervix epidermoid carcinoma cell line, HeLa, human promyelocytic leukemia cell line, HL-60, and rat kidney cells infected with ts25, a T-class mutant of _Rous
sarcoma_ virus Prague strain, _src_ts-NRK, antimicrobial activities against _Staphylococcus aureus_ 209, _Escherichia coli_ HO141, _Aspergillus fumigatus_ Af293, _Pyricularia oryzae_ kita-1,
and _Candida albicans_ JCM1542 and antimalarial activity against _P. falciparum_ 3D7 for 1 were tested. 1 showed moderate cytotoxicity against HL-60 cell lines with IC50 value of 4.4 μg
ml−1. It also showed weak antimalarial activity with IC50 value of 13 μg ml−1, but did not show any effects on other microbes used for the assay up to IC50 values of 30 μg ml−1.
Neoantimycins and antimycins, both of which have a 2-hydroxy-3-formylaminobenzoic acid as a representative functional group, showed potent antimicrobial activity against _C. albicans_,14, 16
and it is reported that the 2-hydroxy-3-formylaminobenzoic acid is essential for the activity.17 However, 1 did not show any effects against _C. albicans_, suggesting that the macrolide
core structure might be also important for the antifungal activity. Compound 1 had the 2-hydroxy-3-formylaminobenzoic acid and phenyl group, which were representative groups for the class of
neoantimycins, and was speculated to have the related biosynthesis pathway with those of neoantimycins. The macrolide core is also the exemplary structure for neoantimycins. However, 1 has
the γ-butyrolactone instead of the macrolide core. Isoneoantimycin is the only metabolite reported as a natural product18 containing above three functional groups without the macrolide core,
but is composed of the identical units with those of neoantimycin. 1 on the other hand does not contain the 2-hydroxy-3-methyl-valeric acid unit found in isoneoantimycin and neoantimycin,
and the hydroxyl group derived from the threonine is dehydrated to form dehydrobutyrine. This type of compound was isolated and reported for the first time as a metabolite from
_Streptomyces_ sp., and analogous metabolites were detected in the fraction library. It is speculated that 1 uses a 2-hydroxy-3-formylaminobenzoic acid as the starter unit using biosynthetic
machinery similar to neoantimycin19 and an unidentified dehydratase catalyzing the formation of dehydrobutyrine. Through the NPPlot screening, we have found some related metabolites to 1,
which show identical UV absorption pattern with slightly different _m_/_z_ values, and will report isolation and structures of these metabolites in the near future. REFERENCES * Osada, H. An
overview on the diversity of actinomycete metabolites. _Actinomycetol._ 15, 11–14 (2001). Article CAS Google Scholar * Newman, D. J. & Cragg, G. M. Natural products as sources of new
drugs from 1981 to 2014. _J. Nat. Prod_. 79, 629–661 (2016). Article CAS Google Scholar * Larsson, J., Gottfries, J., Muresan, S. & Backlund, A. ChemGPS-NP: tuned for navigation in
biologically relevant chemical space. _J. Nat. Prod._ 70, 789–794 (2007). Article CAS Google Scholar * Osada, H. in _Bioprobes_ (ed. Osada, H.) 1–14 (Springer, Berlin, Germany, 2000).. *
Osada, H. in _Protein Targeting with Small Molecules: Chemical Biology Techniques and Applications_ (ed. Osada, H.) 1–10 (Wiley: Hoboken, NJ, USA, 2009).. * Osada, H. & Nogawa, T.
Systematic isolation of microbial metabolites for natural products depository (NPDepo). _Pure Appl. Chem._ 81, 1407–1420 (2012). Google Scholar * Kato, N., Takahashi, S., Nogawa, T., Saito,
T. & Osada, H. Construction of a microbial natural product library for chemical biology studies. _Curr. Opin. Chem. Biol._ 16, 101–108 (2012). Article CAS Google Scholar * Nogawa, T.
_et al_. Verticilactam, a new macrolactam isolated from a microbial metabolite fraction library. _Org. Lett._ 12, 4564–4567 (2010). Article CAS Google Scholar * Nogawa, T. _et al_.
Spirotoamides A and B, novel 6,6-spitoaccetal polyketides isolated from a microbial metabolite fraction library. _J. Antibiot._ 65, 123–128 (2012). Article CAS Google Scholar * Nogawa, T.
_et al_. Pyrrolizilactone, a new pyrrolizidinone metabolite produced by a fungus. _J. Antibiot._ 66, 621–623 (2013). Article CAS Google Scholar * Lim, C. L. _et al_. RK-1355A and B,
novel quinomycin derivatives isolated from a microbial metabolites fraction library based on NPPlot screening. _J. Antibiot._ 67, 323–329 (2014). Article CAS Google Scholar * Lim, C. L.
_et al_. Unantimycin A, a new neoantimycin analog isolated from a microbial metabolite fraction library. _J. Antibiot._ 69, 456–458 (2016). Article CAS Google Scholar * Calglioti, L. _et
al_. The structure of neoantimycin. _Tetrahedron_ 25, 2193–2221 (1969). Article Google Scholar * Takahashi, K., Tsuda, E. & Kurosawa, K. SW-163A and B, novel immunosuppressants
produced by _Streptomyces_ sp. _J. Antibiot._ 54, 867–873 (2001). Article CAS Google Scholar * Ward, D. E., Vázquez, A. & Pedras, M. S. C. Probing host-selective phytotoxicity:
synthesis and biological activity of phomalide, isophomalide, and dihydrophomalide. _J. Org. Chem._ 64, 1657–1666 (1999). Article CAS Google Scholar * Xu, L. Y. _et al_. Antimycins A19
and A20, two new antimycins produced by marine actinomycete _Streptomyces antibioticus_ H74-18. _J. Antibiot._ 64, 661–665 (2011). Article CAS Google Scholar * Izumikawa, M. _et al_.
Novel GRP78 molecular chaperone expression down-regulator JBIR-04 and -05 isolated from _Streptomyces violaceoniger_. _J. Antibiot._ 60, 640–644 (2007). Article CAS Google Scholar *
Takeda, Y. _et al_. Nuclear magnetic resonance and biosynthetic studies of neoantimycin and structure elucidation of isoneoantimycin, a minor metabolite related to neoantimycin. _J. Nat.
Prod._ 61, 978–981 (1998). Article CAS Google Scholar * Li, X. _et al_. Chemical variation from the neoantimycin depsipeptide assembly line. _Bioorg. Med. Chem. Lett._ 23, 5123–5127
(2013). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank Dr Y Hongo in RIKEN for the HRESIMS measurements, Ms H Aono, Ms M Tanaka, Dr J Otaka and Mr K Yamamoto in
RIKEN for activity tests. This work was supported in part by JSPS KAKENHI Grant Numbers 24248022 and 26450148, and grant-in-aid from Research Program on Hepatitis from the Japan Agency for
Medical Research and Development, AMED. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * RIKEN Center for Sustainable Research Science, Chemical Biology Research Group, Wako, Saitama, Japan
Toshihiko Nogawa, Akiko Okano, Chung Liang Lim, Yushi Futamura, Takeshi Shimizu & Hiroyuki Osada * Industrial Biotechnology Research Laboratory, School of Biological Sciences, Universiti
Sains Malaysia, Minden, Penang, Malaysia Chung Liang Lim & Darah Ibrahim * RIKEN Center for Sustainable Research Science, Natural Product Biosynthesis Research Unit, Wako, Saitama,
Japan Shunji Takahashi Authors * Toshihiko Nogawa View author publications You can also search for this author inPubMed Google Scholar * Akiko Okano View author publications You can also
search for this author inPubMed Google Scholar * Chung Liang Lim View author publications You can also search for this author inPubMed Google Scholar * Yushi Futamura View author
publications You can also search for this author inPubMed Google Scholar * Takeshi Shimizu View author publications You can also search for this author inPubMed Google Scholar * Shunji
Takahashi View author publications You can also search for this author inPubMed Google Scholar * Darah Ibrahim View author publications You can also search for this author inPubMed Google
Scholar * Hiroyuki Osada View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Hiroyuki Osada. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information accompanies the paper on The Journal of Antibiotics website SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION (PDF 3608 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Nogawa, T., Okano, A., Lim, C. _et al._ Opantimycin
A, a new metabolite isolated from _Streptomyces_ sp. RK88-1355. _J Antibiot_ 70, 222–225 (2017). https://doi.org/10.1038/ja.2016.113 Download citation * Received: 05 July 2016 * Revised: 02
August 2016 * Accepted: 05 August 2016 * Published: 07 September 2016 * Issue Date: February 2017 * DOI: https://doi.org/10.1038/ja.2016.113 SHARE THIS ARTICLE Anyone you share the following
link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature
SharedIt content-sharing initiative
