Lack of evidence for dna methylation of invader4 retroelements in drosophila and implications for dnmt2-mediated epigenetic regulation
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:

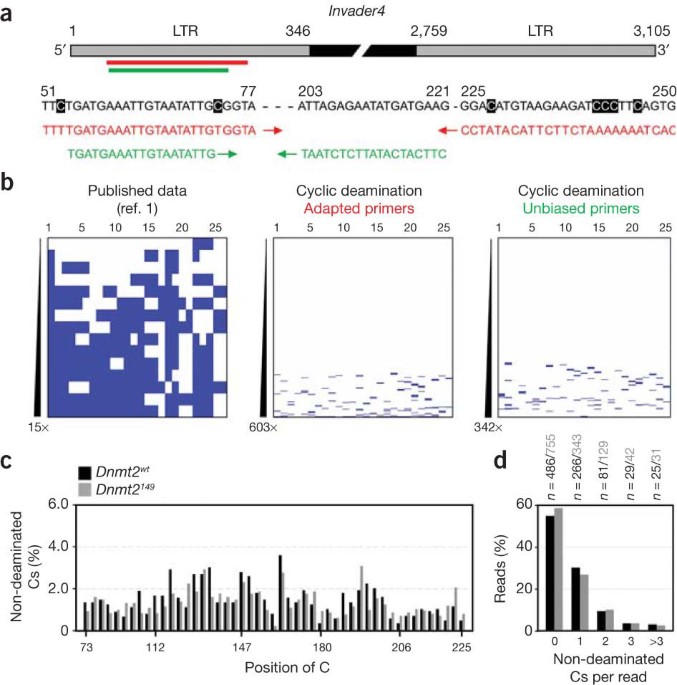
Access through your institution Buy or subscribe To the Editor: In a recent issue of _Nature Genetics_, Phalke _et al_.1 described a new pathway for transposon silencing and telomere
integrity in _Drosophila_ that depends on the DNA methyltransferase homolog Dnmt2. Dnmt2 proteins are the most conserved members of the DNA methyltransferase family, but their substrate
specificity has been discussed controversially within the scientific community2. Previous observations include a weak and highly distributive DNA methyltransferase activity of human DNMT2 in
cell-free assays3, spurious DNA methylation in _Drosophila_4 and highly specific transfer RNA (tRNA) methyltransferase activity for cytosine 38 of tRNAAsp in mice, flies and plants5. All
these observations seemed difficult to reconcile with the processive DNA methyltransferase activity in every dinucleotide context, as observed by Phalke _et al_.1, at _Drosophila Invader4_
retroelements. Furthermore, a recent genome-wide bisulfite sequencing study concluded that transposons are unmethylated in various invertebrates and, specifically, in 0–3-hour-old
_Drosophila_ embryos6. This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues
and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local
taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support ACCESSION CODES ACCESSIONS
GENBANK/EMBL/DDBJ * FBte0000292 REFERENCES * Phalke, S. et al. _Nat. Genet._ 41, 696–702 (2009). Article CAS Google Scholar * Schaefer, M. & Lyko, F. _Chromosoma_ 119, 35–40 (2010).
Article CAS Google Scholar * Hermann, A., Schmitt, S. & Jeltsch, A. _J. Biol. Chem._ 278, 31717–31721 (2003). Article CAS Google Scholar * Lyko, F., Ramsahoye, B.H. & Jaenisch,
R. _Nature_ 408, 538–540 (2000). Article CAS Google Scholar * Goll, M.G. et al. _Science_ 311, 395–398 (2006). Article CAS Google Scholar * Zemach, A., McDaniel, I.E., Silva, P. &
Zilberman, D. _Science_ 328, 916–919 (2010). Article CAS Google Scholar * Schaefer, M., Pollex, T., Hanna, K. & Lyko, F. _Nucleic Acids Res._ 37, e12 (2009). Article Google Scholar
* Schaefer, M. et al. _Genes Dev._ 24, 1590–1595 (2010). Article CAS Google Scholar * Jurkowski, T.P. et al. _RNA_ 14, 1663–1670 (2008). Article CAS Google Scholar * Kuhlmann, M. et
al. _Nucleic Acids Res._ 33, 6405–6417 (2005). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by grants from the Deutsche Forschungsgemeinschaft
to M.S and F.L. (Priority Programme Epigenetics, FOR1082). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Division of Epigenetics, DKFZ-ZMBH Alliance, German Cancer Research Center,
Heidelberg, Germany Matthias Schaefer & Frank Lyko Authors * Matthias Schaefer View author publications You can also search for this author inPubMed Google Scholar * Frank Lyko View
author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.S. performed the DNA methylation analysis. M.S. and F.L. conceived the study and wrote the
manuscript. CORRESPONDING AUTHOR Correspondence to Frank Lyko. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION
SUPPLEMENTARY TABLE, METHODS AND NOTE Supplementary Methods, Supplementary Table 1 and Supplementary Note (PDF 1259 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Schaefer, M., Lyko, F. Lack of evidence for DNA methylation of _Invader4_ retroelements in _Drosophila_ and implications for Dnmt2-mediated epigenetic regulation. _Nat
Genet_ 42, 920–921 (2010). https://doi.org/10.1038/ng1110-920 Download citation * Published: 27 October 2010 * Issue Date: November 2010 * DOI: https://doi.org/10.1038/ng1110-920 SHARE THIS
ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard
Provided by the Springer Nature SharedIt content-sharing initiative
